ShihLab
@LabShih
Followers
1K
Following
612
Media
13
Statuses
451
Twitter of the Lab of Dr. Patrick Shih at UC Berkeley
Berkeley, CA
Joined May 2019
Functional annotations from model organisms are insufficient for robust evolutionary predictions. Active learning across evolutionary space enables generalizable models to accelerate discovery in understudied branches of life. Preprint:
biorxiv.org
Biological discovery and design are increasingly being guided by predictive models in place of costly experimentation. However, existing datasets are often biased by overrepresentation from model...
0
0
0
Our study shows that existing models perform poorly on sequences from non-model organisms because they are out-of-distribution relative to sequences in model species.
1
0
0
Functional characterization of 9,836 candidates and active learning improve model generalizability and provide the first functional annotations for 3,416 proteins from 670 non-model fungi.
1
0
0
We focused on discovery of intrinsically disordered and poorly conserved transcriptional activators across 7.8 million proteins from 2,400 fungal genomes.
1
0
0
Predictive models are increasingly guiding biological discovery since experimentation is time-consuming and resource-intensive. However, existing training datasets are often biased towards model organisms, leading to poor performance in evolutionary studies of non-model species.
1
0
0
Biological diversity beyond model organisms is poorly represented in ML training datasets. Our new preprint uses a high-throughput assay and active learning to discover and characterize gene regulation in non-model organisms.
1
1
1
Big news from Demirer lab: 4 research papers preprinted! Feedback welcomed: eCIS for plant protein delivery: https://t.co/TYKrZNvBWR R2 for targeted insertion: https://t.co/sYyqiriR7e Transient expression: https://t.co/D7UmIeKaMo Chemotaxis assay: https://t.co/poIANSvRsQ 👀⬇️
4
14
72
A truly enabling new approach by Tara Lowensohn, @WillCody_12 and @Sattely_lab to probe plant genetics at scale. Single-gene-per-cell delivery coupled to an effective transcriptional selection system. Libraries in plants. https://t.co/JfLtsLOSjb
0
14
64
Check out our new preprint! We examined sources of variability of transgene expression in N. benthamiana agroinfiltration and ways to minimize it. Graduate student Sophia Tang observed FP expression in >1,800 plants over the course of nearly 3 years!
2
23
51
Causes and consequences of experimental variation in Nicotiana benthamiana transient expression https://t.co/Fa0K5Akado
#biorxiv_plants
0
6
16
Today the Shih Lab (@LabShih) presents ENTRAP-seq, a high-throughput assay for functional screening directly in plant cells.
1
12
44
We're excited to share another work from our lab this week. We present ENTRAP-seq, the first massively parallel reporter assay to characterize protein-coding libraries in plants. Congrats to Simon, Lucas, and all other co-authors. https://t.co/S3gqsCJjp3
biorxiv.org
Transcriptional regulatory domains play key roles in plant growth, development, and environmental responses. Massively parallel reporter assays in yeast and animal cells have enabled large-scale...
0
14
55
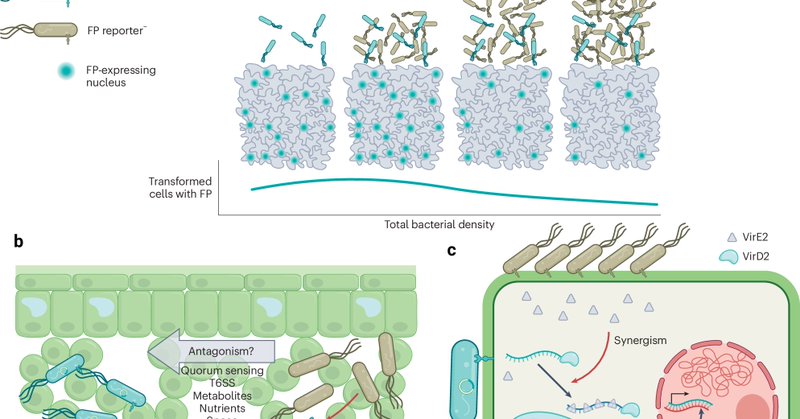
New OA Article: "Quantitative dissection of Agrobacterium T-DNA expression in single plant cells reveals density-dependent synergy and antagonism" https://t.co/qUkht3HYKb News & Views: "Cooperation and antagonism in Agrobacterium-mediated transformation" https://t.co/VQ7MvFQTmI
0
33
112
We also very much appreciate the associated News and Views article:
nature.com
Nature Plants - By integrating theoretical modelling and single-cell analysis of Agrobacterium-mediated transformation in Nicotiana plants, a recent study reveals how agrobacterial density governs...
0
0
6
Happy to share our new publication out today in @NaturePlants where we quantitatively dissect synergy and antagonism that occurs during Nicotiana benthamiana transient agroinfiltration. Congrats to Simon, Matthew, Mitch and all other co-authors! https://t.co/0opNERT3ya
nature.com
Nature Plants - Alamos et al. show that Agrobacterium engages in density-dependent interactions that dictate transformation efficiency and transgene expression. These findings enable a quantitative...
2
35
116
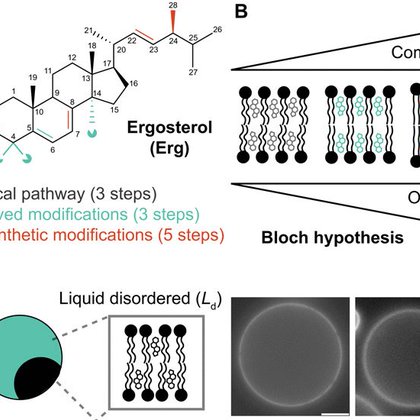
In @ScienceAdvances, we report how small chemical modifications in the metabolism of ergosterol are needed for yeast membranes to phase seperate into ordered yet fluid domains https://t.co/sWDbLiPOFJ (1/n)
science.org
The metabolism of ergosterol shapes this lipid to allow the formation of fluid membrane domains in yeast cells.
7
13
66
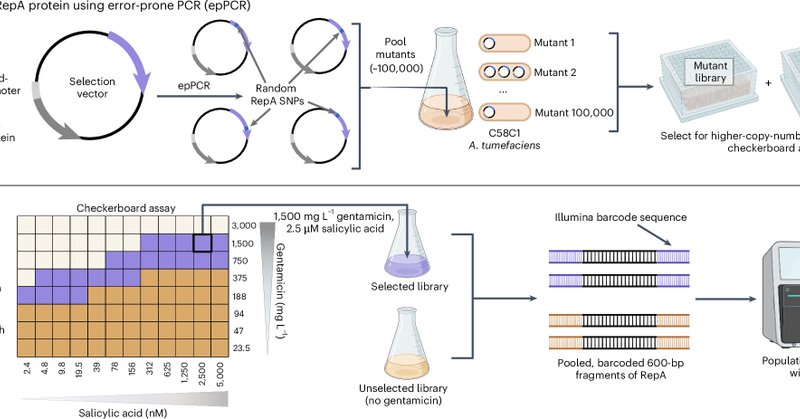
I am pleased to announce new research from the Shih Lab (@LabShih) published today in Nature Biotechnology on improving Agrobacterium-mediated transformation via binary vector copy number engineering ( https://t.co/nCUVQ2ym2p). 🧵A summary thread below🌿
nature.com
Nature Biotechnology - Agrobacterium-mediated transformation of plants and fungi is enhanced by plasmid copy number variants.
5
76
265
Binary vector copy number engineering improves Agrobacterium-mediated transformation https://t.co/ryNgs21Rn5
2
33
89
The Shih Lab is interested in .@UCBerkeley undergraduates who are interested in working with fungi in our lab located at @jbei! If you are interested please DM us!
0
3
2