Ben Williams
@WilliamsSci
Followers
1K
Following
827
Media
19
Statuses
1K
Assistant Professor @PMB_Berkeley. Fascinated by epigenetics, evolution, genomes, and all aspects of plant biology.
Berkeley, California
Joined February 2013
The hidden danger of Biorender (& the death of scientific illustration) A short thread 🧵
41
306
3K
Anthropic is hiring life science researchers. This is very important: applying AI to protein designs, genomics, and advanced cellular simulation... The once thought-to-be 'untreatable' diseases are poised for a reckoning of breakthroughs.
49
92
963
New paper alert! 🔥 By developing engram-specific epigenetic editing tools, our new study provides the first causal evidence that locus-specific epigenetic modifications are necessary and sufficient to drive memory expression. And that irrespective of the memory phase! 🧠🧬
@GraeffJohannes @EPFL Huge congratulations to all authors! and thank you @DrawImpacts for the cover art illustrating how epigenetic modifications act as molecular memory aids!
1
25
118
If you're looking for a postdoc consider this opportunity, it is for a *really* cool NSF-funded project:
0
1
5
We are hiring! If you have experience in plant genomics + AI, or promoter engineering + synthetic biology, check out our two open postdoc positions (computational + wetlab): https://t.co/pdtXE8gYAN please RT #ERC
1
20
31
Our new work uncovering how WIND1 promotes somatic embryogenesis as a bifunctional chromatin regulator!!! We knew WIND1 promotes new fate acquisition and now we show WIND1 also helps repressing existing identity!
WIND1 controls cell fate transition through histone acetylation and deacetylation during somatic embryogenesis https://t.co/2K8lcfrBBG
#biorxiv_plants
1
17
48
The Todesco lab at MSL UBC is looking for a Research Tech to help us run the lab and do fancy science! Good wet lab skills and enthusiasm for plant research are required, experience with genomics and bioinformatics is a plus. Spread the voice! 🧬🌻🧪🌱 🧫 https://t.co/N2VhP7Vmw2
0
8
18
Few things in science are as heart-warming as seeing one of your PhD or postdoc buddies start their own lab and begin absolutely KILLING IT with amazing discoveries!
I am excited to share the latest paper from my lab where we leverage the selection bias of condensate-promoting oncofusions to uncover molecular rules governing condensate specificity and function. https://t.co/AQlErs3lni.
1
0
6
I am excited to share the latest paper from my lab where we leverage the selection bias of condensate-promoting oncofusions to uncover molecular rules governing condensate specificity and function. https://t.co/AQlErs3lni.
11
39
202
please share - 2 postdoc positions available - molecular physiology of stomata and plant water use of efficiency https://t.co/QzgbGKEzQy
@RIPEproject @ASPB @Stomata_Tweets @PlantPhys
0
18
29
Delighted to share this. A novel H4 variant that modulates chromatin to mediate salt stress response. This mammoth work was driven to perfection by @VivekHariSunda1, a wonderful PhD student who also taught me. We report its CryoEM structure & unusual chemistry @NCBS_Bangalore 1/3
New Article: "An Oryza-specific histone H4 variant predisposes H4 lysine 5 acetylation to modulate salt stress responses" https://t.co/e4usthp5xG An Oryza-specific histone H4 variant (H4.V) forms condensed, less stable nucleosomes, regulating the salt stress transcriptome.
34
29
146
> bacteria were engineered to make molecules that absorb light in unique ways. > the microbes were buried in soil. > using a drone with a hyperspectral camera, one can "see" where the microbes are buried from ~300 feet away. one of the most sci-fi papers I've seen in awhile.
12
137
815
In 2004, it took the world a year to add one gigawatt of solar power — now it takes a day.
79
607
2K
Extremely clever new NGS tech from Roche 🧬 If it's hard to discriminate between nucleic acids accurately with a nanopore, why not synthesize a new polymer off a DNA template that is easier to sequence? It's an intuitively simple idea, but took *a ton* of creative nucleic acid
16
192
910
Transcription factors instruct DNA methylation patterns in plant reproductive tissues https://t.co/02iI5jAeEx
#biorxiv_plants
biorxiv.org
DNA methylation is maintained by forming self-reinforcing connections with other repressive chromatin modifications, resulting in stably silenced genes and transposons. However, these mechanisms fail...
0
13
19
Excited to share work from Zhongshou Wu in our lab showing that GDE1 and REM transcription factors help shape DNA methylation patterning. Together with work from Julie Law’s lab also just out, this shows an important way that genetics directs epigenetics.
biorxiv.org
DNA methylation plays crucial roles in gene regulation and transposon silencing. In plants, the maintenance of DNA methylation is controlled by several self-reinforcing loops involving histone...
0
18
47
🌱 Egg cell-specific degradation of CENH3 using engineered E3-ligases successfully induce haploids ⚠️ Preprint from Andreas Houben's lab @LeibnizIPK 👉🏼 Gametophytic degradation of CENH3 - a synthetic biology approach for haploid induction in plants 📖 https://t.co/MRKivuEsYA
1
27
83
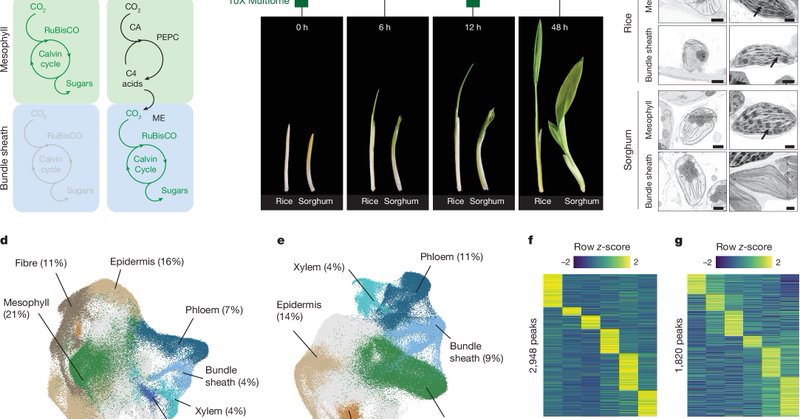
Very pleased to see this work out that identifies a code in C3 rice co-opted to allow C4 photosynthesis. https://t.co/CNuFLMr13J Thanks to all involved but in particular @l_luginbuehl @JoeEcker @_joseph_swift Great transatlantic partnership. @plantsci
nature.com
Nature - Single-nucleus RNA-sequencing and chromatin-accessibility analyses in rice (a C3 plant) and sorghum (a C4 plant) provide insight into how C4 photosynthesis evolved in bundle-sheath cells,...
7
42
165
Here is something to distract you from the gloom: What can metagenomics tell us about the taxonomic composition of lichen symbioses? A peer-reviewed version of our analysis of >400 lichen metagenomes from around the world is now out in @PLOSBiology. 1/12 https://t.co/MMGDSEwm8H
5
57
154