Andy Dahl
@andywdahl
Followers
583
Following
837
Media
0
Statuses
52
Assistant professor at uchicago working on statistical genetics methods
Joined June 2019
Monotone convex transformations of an additive phenotype induce sign-consistent GxE--new preprint from @sadowskimich, Noah Zaitlen and Richard Border
New preprint. We show that a wide class of outcome transformations and endogenous treatment effects induce sign-consistent gene-environment interaction (GxE) effects. https://t.co/YwjjiQ2kL0 1/4
0
3
9
I'm thrilled that my lab at NYU is now supported by an NIH MIRA grant! I'm looking to hire 1-2 senior lab members (outstanding postdoc candidates or experienced staff scientists) with expertise in computational and statistical methods in human genetics or genomics. Please share!
5
64
177
Very happy to share our new paper now on @medrxivpreprint: “Genetic risk effects act in sets”, a great effort led my PhD student @JolienRietkerk, and performed together with collaborators @andywdahl, @AndrewSchork, @jonathan_flint1 etc. Thread 1/n
2
34
104
🚨 Applications are open for the UCLA Postdoct Training Program in Neurobehavioral Genetics 🧠🧬 Deadline: 6/30 👉 https://t.co/k8nPEs4Vs1 Contact me if you're interested to write a proposal together combining genetics, functional genomics, & digital phenotypes from smartphones!
1
6
7
Congratulations to Joel on the publication of this paper in AJHG. He shows that polygenic scores derived from different quantiles of a phenotype differ, and uses this to come up with an interesting and novel test for confounds and GxE https://t.co/FQYD8GI9zh
0
12
34
Are you an early-career researcher working on the computational analysis of population-level human genetics data? We want to hear from you about if, how, and why you use population descriptors in your research! Fill out our short survey: https://t.co/7K4Ro23Bsq
0
4
4
Very happy to share *two* new projects from my group now on @biorxivpreprint on mtDNA variation effects on tissue-specific gene expression conducted using RNAseq data in 49 tissues in the @GTExPortal project! 🧵 1/n
2
11
55
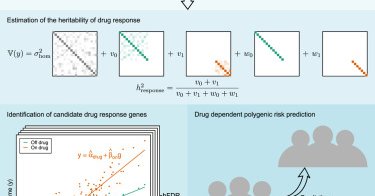
GxDrug interactions now published at cell genomics :) @sadowskimich did a ton of great work in this paper. We hope the approach is a step toward improving predictions for individual-specific treatment response
Our work using gene-environment interaction methods to study the genetics of drug response is now out in @CellGenomics. @CellPressNews
https://t.co/slbAWPLvKG
0
7
26
Our work using gene-environment interaction methods to study the genetics of drug response is now out in @CellGenomics. @CellPressNews
https://t.co/slbAWPLvKG
cell.com
Sadowski et al. propose a framework to study the genetics of response to commonly prescribed drugs in large biobanks. They quantify the heritability of response to statins, metformin, warfarin, and...
0
15
54
@CompMedUCLA postdoc Richard Border is moving to Carnegie Mellon in Spring 2025 his own lab as an assistant professor in the CMU Department of Computational Biology. @CMUCompBio @SCSatCMU
https://t.co/8L2jGHLkDI
1
7
22
Our new work uses GxDrug interaction to model genetic effects on drug response in cross sectional data. Includes a new method for GxE TWAS. There are important caveats, but we hope this is a step toward more precise treatments
New preprint. In this paper we quantify heritability of drug response and identify involved genes by applying gene-environment interaction methods to biobank data. https://t.co/mXvNhg8ksY 1/4
0
9
28
Our study on utilizing smartphones to track behavior in real-time & predict depression severity is now out in Nature Digital Medicine. @jonathan_flint1 @sr_sankararaman @halperineran @UCLAThinkGrand
1 in 5 grapple with #depression, making it a leading cause of global disability. This study utilizes smartphone sensors to track behavior in real time & predict depression severity up to 3 weeks ahead, offering possibilities for personalized treatments. https://t.co/jLGVTVk3e1
0
14
36
Open TT faculty position at the department of medicine at the University of Chicago! Come join us in Genetic Medicine, Biomedical data science, Oncology, or any of the other sections. Apply here:
0
24
30
Great summary. Thank you Sasha :)
Really cool paper that investigates (primarily mental health) phenotype imputation for GWAS as a trade-off between increased power versus decreased specificity. This gets at the broader conceptual question of what is a phenotype for? A few intriguing results/directions. 🧵:
0
0
4
New paper by @andywdahl and I on phenotype integration improving GWAS power while preserving specificity is now out in @NatureGenet 🎉 thank you all collaborators and reviewers!
7
63
246
Really cool paper that investigates (primarily mental health) phenotype imputation for GWAS as a trade-off between increased power versus decreased specificity. This gets at the broader conceptual question of what is a phenotype for? A few intriguing results/directions. 🧵:
New paper by @andywdahl and I on phenotype integration improving GWAS power while preserving specificity is now out in @NatureGenet 🎉 thank you all collaborators and reviewers!
2
10
57
Less than a week to #wcpg2023, looking forward to seeing everyone there! My lab is hiring postdocs to work on quantgen methods, EHR utilisation, disease heterogeneity and trajectories, if interested please DM, also happy to chat during the conference! https://t.co/VQgmoq2cmq
sites.google.com
Cai Lab Cai lab is based in the Department of Biosystems and Engineering (D-BSSE) in ETH Zürich and the Basel Research Centre for Child Health (BRCCH). We are quantitative geneticists working on...
2
13
33
Excited for the 2nd annual PhD Info session with @UChicagoGGSB Sign up at https://t.co/emtIRs4BxJ for application tips from a diverse panel of current PhD students and faculty! @QuestBridge @ImFirstGen @LEDA_Scholars @TheJKCF @ReachHigher @FirstgenCenter @HSFNews @cokescholars
0
10
6
The Department of Human Genetics at University of Chicago is searching for a faculty candidate in Computational genetic and genomics. Please share this job ad with your colleagues:
0
71
81
Very happy to see Minhui's paper out :) It was WAY more technically challenging than expected, scRNA data is complicated... But Minhui was up to the challenge, and we hope this is a step toward robust Gaussian variance component models for scRNA
New preprint! Together with @andywdahl , we develop CTMM (Cell Type-specific linear Mixed Model), a novel method for scRNA that detects cell type-specific gene expression from a new angle compared to standard tests for Differentiatlly Expressed Gene (DEG).
2
2
33