Minhui Chen
@chen_cmh
Followers
372
Following
1K
Media
10
Statuses
218
Assistant Professor in Statistical Population Genetics @VCU_VIPBG | Former Postdoc at @UChicagoMed @usc_cge.
Joined July 2015
Great science happens at the intersections. STATGEN is a fantastic forum to connect, challenge assumptions, and raise the bar for rigor in genetics and genomics. Submit an abstract by December 15, 2025: https://t.co/yFHKXwqV2U Conference Website:
docs.google.com
STATGEN 2026 (https://statgen26.emory.edu) will be held at Emory University in Atlanta Georgia May 18-20, 2026 . Please use this form to submit a title and an abstract by December 15, 2026 to be...
0
8
26
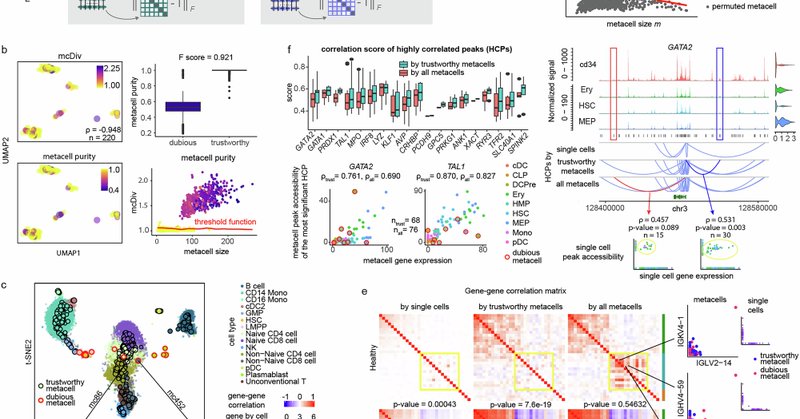
1/3 Metacells boost power in single-cell RNA-seq & multiome analysis. But without checking homogeneity, they risk forming dubious metacells that bias discoveries. We introduce mcRigor: a statistical safeguard for rigorous metacell analysis. 👉
nature.com
Nature Communications - Aggregating similar single cells into metacells is a common heuristic for sparse data, but risks mixing dissimilar cells. Here, authors present mcRigor, which detects and...
3
34
144
🚀 Introducing Paper2Agent - a system that turns static research papers into interactive AI agents you can chat with and use the tools/data in the paper. Just ask, “Run this paper’s method on my data.” The paper agent will handle the task for you. 1/n
17
92
502
excited to share our new preprint looking at mosaic chromosomal alterations in blood whole genome sequencing data! i learned tons working on this project, and i hope our findings are of interest to those thinking about CH, somatic mosaicism, and genetics. https://t.co/HUgGESfBH8
medrxiv.org
Clonal expansions of hematopoietic cells carrying mosaic chromosomal alterations (mCAs) are commonly detectable in elderly individuals. Here, we studied 43,617 autosomal mCAs that we ascertained in...
3
13
42
Very happy to share our new paper now on @medrxivpreprint: “Genetic risk effects act in sets”, a great effort led my PhD student @JolienRietkerk, and performed together with collaborators @andywdahl, @AndrewSchork, @jonathan_flint1 etc. Thread 1/n
2
34
104
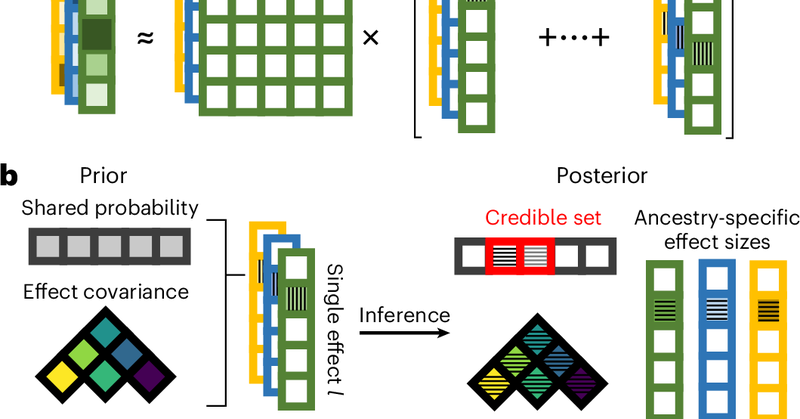
Happy to share that our work from the @nmancuso_ lab is out in @NatureGenet! We developed SuShiE, a multiancestry fine-mapping method for molecular traits.
nature.com
Nature Genetics - SuShiE is a multiancestry fine-mapping method for molecular quantitative trait loci that leverages linkage disequilibrium heterogeneity to improve resolution, infer cross-ancestry...
3
36
98
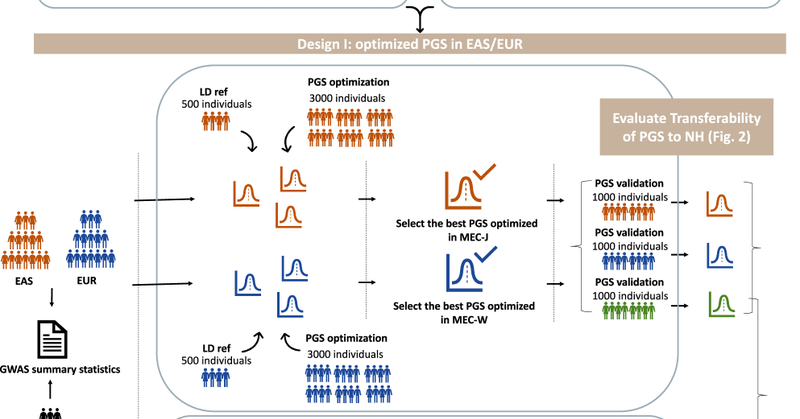
The fifth paper from the group this year. Led by a former postdoc @imyingchulo, our evaluation of polygenic scores (PGS) for BMI and T2D for Native Hawaiians is published in @CommsBio! https://t.co/LpuPI02KvG
nature.com
Communications Biology - An evaluation of polygenic score prediction models for BMI, height, and type-2 diabetes in Native Hawaiians revealed that these models tend to underperform for Native...
1
3
3
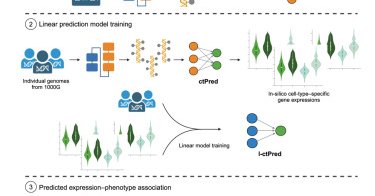
Check out our scPrediXcan paper https://t.co/Pcni6LfrNg Led by talented @Charles_Zhou12, supervised by @MengjieChen6 and me, with thanks to many contributors scPrediXcan integrates deep learning and single cell expression data into a powerful cell type specific TWAS framework.
cell.com
Zhou et al. introduce scPrediXcan, a novel transcriptome-wide association study framework that integrates the deep learning-based model ctPred for cell-type-specific expression prediction. Applied to...
0
23
63
Work with @jnovembre on extending FEEMS to represent long-range gene flow on a background migration surface! The software for the method, called FEEMSmix, is readily available at https://t.co/SsOeAc2c7c
#popgen #Genetics #evolution
github.com
Forked repo with an initial goal to add features such as long-range migration. - GitHub - VivaswatS/feems: Forked repo with an initial goal to add features such as long-range migration.
Jointly representing long-range genetic similarity and spatially heterogeneous isolation-by-distance https://t.co/j5usRlJsjm
#biorxiv_evobio
1
15
40
Family members share quite a lot genetically and environmentally. Curious about how we quantify the resemblance using any possible genealogy-traceable relatives to study human phenotypes?Join us at Exhibit hall Board 4118w to explore our latest work! 😀 #ASHG24
0
4
13
Excited to be heading to #ASHG24 today! I’ll be presenting at Wednesday’s poster session (Board 3016W) about our latest work on spatial sampling strategies & deleterious variants - come say hi!
1
3
17
I am very excited to share my study, trans-regulation of proteome through protein-protein interactions, at #ASHG2024 ! Please check my poster number 6077 on Wednesday (11/6) afternoon!
0
3
26
Our work on trans regulation of proteome and how protein-protein interactions play a role in it🧬.
🚨New Preprint Alert: Our work led by @JinghuiLi14637 is up on bioRxiv. We re-analyzed large maps of trans-pQTLs, and found that protein–protein interactions shape trans-regulatory impact of genetic variation on protein expression and complex traits. A brief thread🧵👇
0
2
12
Excited to share our work on alternative splicing (AS) https://t.co/9zf3WRSPR5. This version, compared to the biorXiv one, is much more succinct due to word limits; I hope you will have a look. Congrats to @BenjaminFair3 and @cfbuenabad for their incredible work!🧵⬇️
5
33
106
My lab continues to explore questions on AS, unproductive splicing, and co/post-txnal gene regulation. I would love to hear from you if you might be interested in joining us — we are a small but well funded group for experimental and computational work. https://t.co/Vc2oCkhD7D /n
1
2
5
Please RT!
🧬Postdoc Opportunity in Statistical/Computational Genetics!🧬 Join our team in Human Genetics @PittPubHealth! Focus: Genetic etiology of cardiometabolic disease in Samoans & Pacific Islanders. Collaboration with @CharlestonCWKC
https://t.co/GH1Bpgb8qG
#Postdoc #Academicjobs
0
2
2
Excited to share the last paper from my PhD, advised by @spence_jeffrey_ and @jkpritch! We explore a new way to study selection on complex traits that circumvents some of the most troublesome limitations of GWAS https://t.co/bGFtmwz2ia (1/9)
biorxiv.org
Natural selection on complex traits is difficult to study in part due to the ascertainment inherent to genome-wide association studies (GWAS). The power to detect a trait-associated variant in GWAS...
2
40
109
New work about the Finnish pedigree and autoimmune diseases from @feiyi_wang, co-supervised by me, @andganna, and @TinskuT! Preprint:
medrxiv.org
Type 1 diabetes (T1D) and other autoimmune diseases (AIDs) co-occur in families. We studied the aggregation of 50 parental AIDs with T1D in offspring and the contribution of a shared genetic backgr...
T1D and other autoimmune diseases (AIDs) co-occur in families. How parental AIDs impact T1D risk in offspring and how much of the familial risk is explained by HLA and non-HLA variants? 👩🏻🦱🧔🏻♂️👶🏻 Find out more from this thread 🧵 and our new preprint ⬇️! https://t.co/CWCcvd9VR2
0
2
13
So proud that this work is now published @AJHGNews. It was truly an effort of a team of undergrad students (at the time), led by @itscahoon. It was also a project used to introduce new grad students or others just getting into genomic analysis - the team effort really worked out!
📢New from @itscahoon @CharlestonCWKC & co 📰Imputation accuracy across global human populations https://t.co/VcrmSLCTTv
1
11
30