John Lovell
@_johntlovell
Followers
869
Following
745
Media
29
Statuses
241
Evolutionary biologist and outdoor enthusiast | comparative & quantitative genomics (mostly plants) | HudsonAlpha Genome Sequencing Center & Joint Genome Inst.
Joined February 2021
Link to applications and details here:. Postdoctoral Research Associate - Plant Systems Biology & The Center for Bioenergy Innovation (CBI) Job Details | Oak Ridge National Laboratory.
jobs.ornl.gov
Postdoctoral Research Associate - Plant Systems Biology & The Center for Bioenergy Innovation (CBI)
0
0
2
🚨 We're looking for a #postdoc to join our team at the Center for Bioenergy Innovation at Oak Ridge National Labs, TN 🚨. Project: Advance poplar bioenergy by understanding adaptive trait/genomic variation using breeding, pop gen, and bioinformatics. Apply by 18-Aug.
1
2
4
Wow, the cover looks great! .Nice work Patrice and @roederlab.bsky.social. The GENESPACE plot uses our new @jgi Pennycress and Brassica rapa genomes built in collaboration with @SpicyBotrytis & @ktgreenham, hosted on @phytozome.
Check out the beautiful cover to our focus issue on Translational research from Arabidopsis to crop plants and beyond. More articles coming shortly. Congrats Patrice Salome and @_johntlovell
0
0
3
Happy to see this out!! Check out my thread over at the better place for some quick background and summaries.
Improving American chestnut resistance to two invasive pathogens through genome-enabled breeding #biorxiv_genomic.
0
0
3
RT @aeharkess: We are still looking for speakers for the PAG Plant Reproductive Genomics session - please reach out if interested!.
0
14
0
RT @AdamRutherford: Ok, here we go: Much of my work concerns the history and return of scientific racism. I’ve written extensively about at….
0
2K
0
RT @carriewessi: Join us!! I'm recruiting a postdoc and grad student to work with us on parallel trait evolution, quant and pop gen, and sp….
0
84
0
RT @zevkronenberg: Very excited to share our work on building a benchmark of all classes of variation based on the large CEPH-1463 pedigree….
biorxiv.org
Recent advances in genome sequencing have improved variant calling in complex regions of the human genome. However, it is difficult to quantify variant calling performance since existing standards...
0
28
0
There are so many things to like about this paper — very cool data viz, nuanced treatment of complex plant #pangenomes, integrations with traits. I also appreciate a lack of the non-sensical term "super pan-genome" that is somehow now in vogue😉.
🚨 Excited to share this #preprint from my postdoc in the Lippman lab @CSHL, in collab. with @mike_schatz & many others! Using pan-genomics & pan-genetics across the Solanum genus 🍅🥔🍆 we reveal gene duplications 🧬 as contingencies in crop engineering.
0
0
6
Love this!!! This is why we are calling the assembly-only follow up “DEEPSPACE”.
Every time with my favorite GENESPACE pipeline by @_johntlovell . (those who know can tell if the shooting stars here are allo or auto 😉)
0
0
9
RT @njkooyers: Our preprint is up! Using a pop genomic dataset spanning 6 continents and 4 common gardens in the native & invasive range o….
biorxiv.org
Rapid adaptation during invasion has historically been considered limited and unpredictable. We leverage whole-genome sequencing of >2600 plants across six continents to investigate the relative...
0
15
0
2
0
1
I'm putting together an analysis of variation between genomes in model systems . what are the *two* most-used reference genomes for the following?.Human -- Hg38 / CHM13.Mouse -- GRCm39 / ?.Arabidopsis -- Araport11 / ?.Maize -- B73 / ?.Rice -- (?IRGSP1?) / ?.Soybean -- Wm82 / ?.
4
1
6
RT @KodaniLab: The @KodaniLab is looking for a talented lab manager and senior scientist. Come join us @hudsonalpha and work alongside @nnu….
0
6
0
All this work was led by @AdamHealey65, and made possible with funding and support from @hudsonalpha @jgi @BerkeleyLab @jbei. Find the genome on @phytozome:
1
0
0
So, back to the original question - how can we make a sugarcane genome that is useful? To improve sugarcane, breeders look for genetic diversity within individuals; these are sites that segregate in selfed progeny populations — we need a genome that represents this diversity.
1
0
0
However, in sugarcane this isn't appropriate: since gene copy number varies we cannot know the physical location of identical sequences. This means we cannot and should not produce a 'T2T' assembly for every all 120 chromosome in R570.
1
0
0
In simpler genomes like auto-tetraploid potato, IDB blocks due to inbreeding can be computationally duplicated so that each biological haplotype has the full complement of sequence - a 'T2T' assembly.
1
0
0
But, it gets more complex. As the pedigree above shows, there is lots of inbreeding in hybrid sugarcane (R570 has only three grandparents), and about half of domesticated haplotypes are identical by descent (IBD) within the last century.
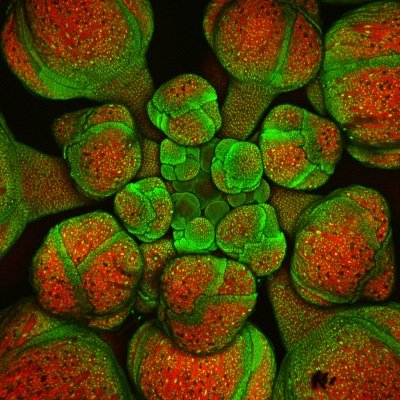
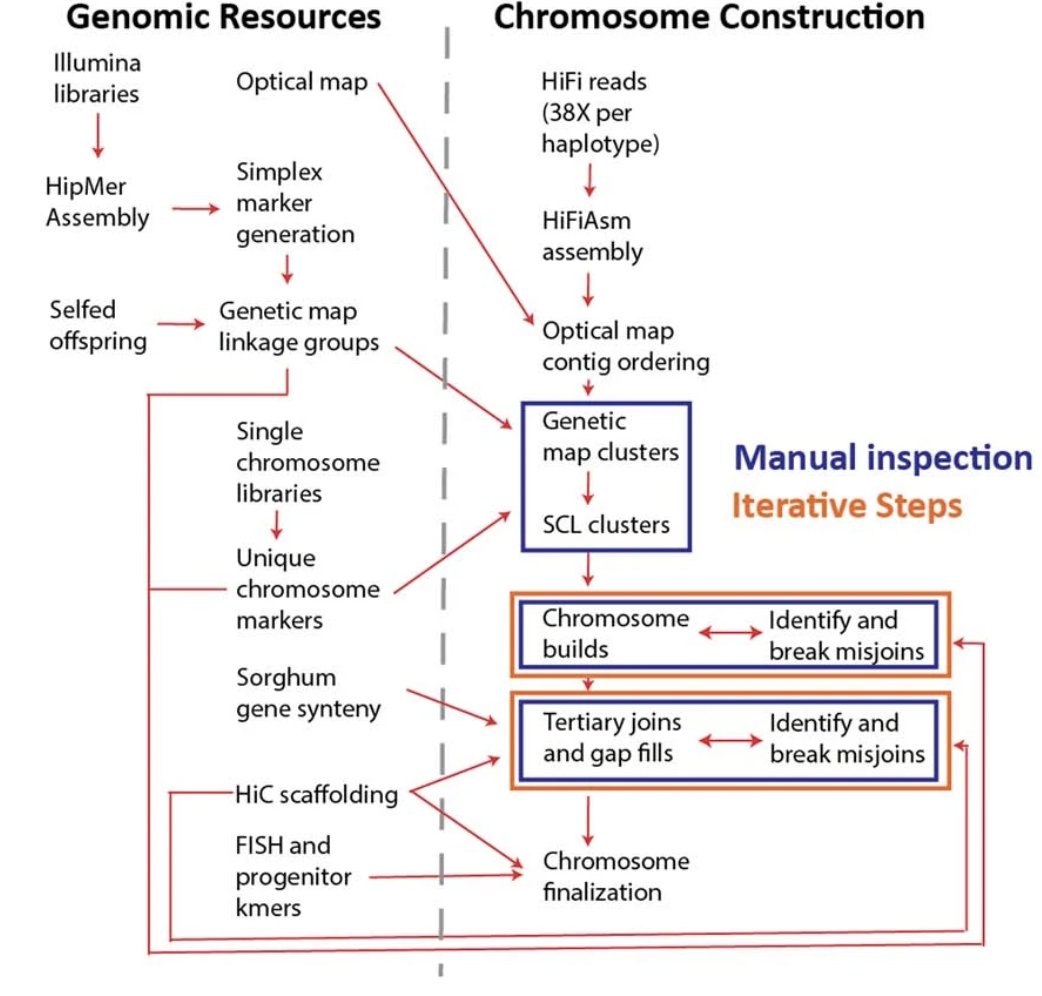
It's finally published! The first polyploid reference genome for sugarcane cultivars. This is the most complex plant genome we have ever completed and a fantastic resource for sugarcane breeders and researchers! @hudsonalpha @jgi @BerkeleyLab @jbei.
1
0
0