Thomas Welte
@WelteThomas
Followers
78
Following
94
Media
3
Statuses
71
Clinician scientist @unispital_usz, @UZH_en, @SFB1453_NephGen @NCCR_RNADisease. RNA + dev. biology.
Joined August 2019
Novel insights into the silencing of an understudied transposable element - check out our latest preprint by Jakob Schnabl-Baumgartner and colleagues https://t.co/sQUYsJQjK1
biorxiv.org
Short interspersed nuclear elements (SINEs) are abundant non-autonomous transposable elements derived from RNA polymerase III (POL III)-transcribed short non-coding RNAs. SINEs retain sequence...
0
6
17
Welcome to @WelteThomas as a new Associate Member of the NCCR RNA & Disease! Clinician Scientist at University Hospital Freiburg and @FMIscience . Soon establishing a research lab on post-transcriptional gene regulation in organ development and repair at the @UZH_Science.
0
2
5
Glad you are still here. Let others know and RT: https://t.co/6Ayq56C9oC
🚨Preprint alert! We reveal how selective #autophagy of ribosomes balances survival vs growth in starving #Celegans! Read it here: https://t.co/c2qcwbE28r. Led by fantastic @Joelfornow with @DengjelL @nstroustrup1 from @IZB_unibern @unibern @unifr @CRGenomica 🧵
0
1
0
Excellent choice - congratulations, Victor and Gary, for this deserved recognition of your breakthrough work!
BREAKING NEWS The 2024 #NobelPrize in Physiology or Medicine has been awarded to Victor Ambros and Gary Ruvkun for the discovery of microRNA and its role in post-transcriptional gene regulation.
0
4
36
Tissue-resident memory T cells break tolerance to renal autoantigens and orchestrate immune-mediated nephritis
pubmed.ncbi.nlm.nih.gov
Immune-mediated nephritis is a leading cause of acute kidney injury and chronic kidney disease. While the role of B cells and antibodies has been extensively investigated in the past, the advent of...
0
0
2
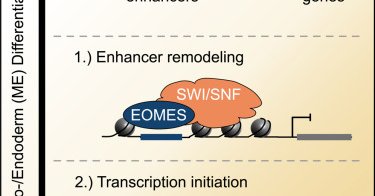
A novel protein complex, ChAHP2, is involved in silencing of ERVs and LINE1 elements, complementing the SINE-targeting ChAHP complex, to bring about efficient retrotransposon repression. @MarcBhler4 @JosipAhel Learn more here: ➡️ https://t.co/ekqtQL0l14
0
11
22
Preprint by @LabGrosshans (@FMIscience) & @ward_lab (@ucsc) w/ 1st authors Rebecca Spangler, @GuinevereAshley, @KathrinBraun16 "A conserved chronobiological complex times C. elegans development" @biorxivpreprint
https://t.co/VjGUTJRVs7
0
3
6
Two more weeks to apply for #PhD position in #Quantitative #Developmental #Biology using #Celegans. Join us to study how growth signals control development and aging in space and time. GIF is shows bursts of FoxO/DAF-16 activity. https://t.co/aKEAxSaXEj
Thrilled to see images from #SQUID micropscopes coming in: 1 week in the life of a #Celegans. @PrakashLab @hongquan_li @cephlainc. Join us for a #PhD if you are as excited about Growth and Aging as we are: https://t.co/YprW2cmvBE
0
7
9
What's better than publishing a paper? Publishing it next to another paper from your former postdoc's lab. Congratulations, @mdlmata & Team! Full stories on how miRNAs get degraded at https://t.co/nzkiIkz7aq & https://t.co/aUO9MoBibw
@NAR_Open @FMIscience
1
7
27
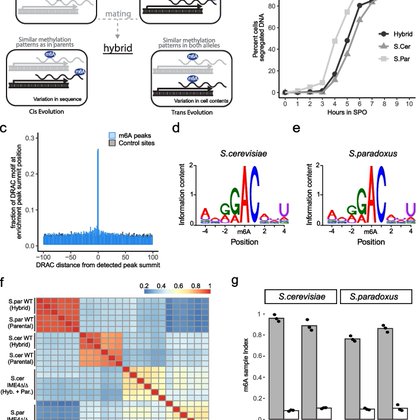
What are the forces shaping m6A evolution? Using interspecies hybrids in yeast and mammalian systems, we find that changes in m6A levels are nearly exclusively driven in cis.
genomebiology.biomedcentral.com
Background N6-methyladenosine (m6A) is the most abundant mRNA modification, and controls mRNA stability. m6A distribution varies considerably between and within species. Yet, it is unclear to what...
2
30
116
Ever wondered how biological clocks time developmental events? @LabGrosshans @FMIscience we do - and use experiments and theory to find out. Join us – FMI's PhD program is now accepting applications 👇, and we offer #phdpositions.
📢We're now accepting applications for our #PhD and #MDPhD programs! 🧬Join us in Basel, Europe's top hub for the #lifesciences, and unleash your potential in a vibrant, international and supportive environment 📅Apply by May 1 at https://t.co/NSIgFeL5A3
#AcademicTwitter
0
7
3
Congratulations Luke!
@mscienceaustnz is proud to once again sponsor the Mid-Career Awards at @GenomeConf. Congratulations to @LukeIsbel and @HaloomRafehi. Impressive research and excellent communication.
1
0
2
@PetersLabFMI: Excited to share our latest preprint lead by Greg Fanourgakis (@Grigo_Fanou). Key findings are that DNA methylation modulates nucleosome retention in sperm and H3K4 methylation deposition in early mouse embryos.
biorxiv.org
DNA methylation (DNAme) serves a stable gene regulatory function in somatic cells ([1][1]). In the germ line and during early embryogenesis, however, DNAme undergoes global erasure and re-establish...
2
17
55
We’re thrilled to report the discovery of ChAHP2! Its chromatin binding specificity is distinct from ChAHP, predominantly associating with ERV and LINE1 retrotransposons via HP1beta-mediated binding of H3K9me3.
biorxiv.org
Retrotransposon control in mammals is an intricate process that is effectuated by a broad network of chromatin regulatory pathways. We previously discovered ChAHP, a protein complex with repressive...
3
22
92
Our paper is out today in @Nature! Imagine getting this plot when studying single-cell data of growing bacteria! Could it be the cell cycle?! We found that a gene’s response to its replication reveals gene regulation on a genomic scale. https://t.co/KQidVr6WKZ
@AndrewPountain
38
229
1K
In our new paper in @NatureComms ( https://t.co/ie7C58WFKr) we show #robustness of pharynx-to-body #scaling to tissue-specific growth inhibition involving the mechanotransducer #YAP-1 https://t.co/ie7C58WFKr. 👇for 🧵. PhD positions available for follow-up (Pls RT). @IZB_unibern
Excited to share our new preprint on #organ #growth in #Celegans
https://t.co/06anEi02JV. Led by fantastic @klemento6 with @themairlab @IZB_unibern @unibern @HarvardChanSPH we showed how ultrasensitive coupling of body and pharynx growth by #YAP ensures pharynx size uniformity🧵
1
8
31
Massively parallel mapping of substrate cleavage sites defines dipeptidyl peptidase four subsite cooperativity
0
0
3
Great collaboration by @SFB1453_NephGen scientists Frederic Arnold and @LukasWesterman4
#diabetes #KidneyWeek2023
0
0
1
How does sugar consumption affect renal health? Data from ~19k patients at @Uniklinik_Fr show the scary truth: Renal risk starts to increase already in the prediabetic range. Find out more in our newest publication in @JInternMed: https://t.co/zOFgdwQGYo
2
3
7