Thomas Miller
@TCR_Miller
Followers
841
Following
377
Media
15
Statuses
195
Associate Professor at the University of Copenhagen. Interested in determining the molecular mechanisms of eukaryotic genome replication using cryo-EM.
Copenhagen, Denmark
Joined August 2013
I am happy to announce that we've recently posted our new manuscript, titled: ‘Cell Cycle Regulation has Shaped Budding Yeast Replication Origin Structure and Function’ on BioRxiv ( https://t.co/E9siBoXi36). We hope you enjoy! Tweetorial below:
4
13
50
I have received a Wellcome Discovery Award to study the initiation of eukaryotic DNA replication. 8 years of funding at the Crick Institute. Looking for a postdoc in molecular mechanism, cryo-EM and in situ structure? Contact me now! 3 postdoc positions advertised soon.
6
19
93
🚨Hiring a Research Technician!🚨 Join our lab to study mobile DNA and its impact on biology and genome engineering! 🌟 MSc. in Molecular Biology, Biochemistry or related field required. PhD highly desired. 🌟 70% Research 30% Admin 🌍 @MaxPerutzLabs, @viennabiocenter #Austria
2
62
143
This is a great opportunity! Come join Kathleen in Copenhagen!
We're hiring! We have a funded postdoc available and encourage anyone interested in epigenetics and/ or gametogenesis to apply. Experience in mouse genetics/ development especially welcome! Deadline: 1 October Apply here: https://t.co/yWYqQt5YRs Please retweet!
0
0
1
Exciting news! I am grateful and thrilled to join the Section for Biomolecular Sciences #UCPH. We use single-molecule tools to study how pioneer factors interact with chromatin and promote cell reprogramming. Join our ERC-project PIONEER, now recruiting on all levels!⬇️
5
21
63
Congratulations all! Well deserved!
A huge congratulations 🙌 to @groth_anja, @ChoudharyLabCPR, and @duxin_lab on these fantastic and well-deserved achievements with @ERC_Research grants🏆🍾 #CPRlegacy
0
0
7
Are you interested in DNA replication, DNA repair or chromatin? Do you have a passion to find out how proteins function in these processes? Are you collaborative and ambitious? Check out these open positions in my lab (and @FMattiroli lab):
hubrecht.eu
1
28
45
🚨 Join us! We're seeking a Lecturer in Cell & Molecular Bioscience at UOW. Leverage our world-class cryo-electron microscopy facility for pioneering research in structural biology. Apply now! Any questions reach out!
0
10
11
Our new paper is out today in @ScienceMagazine showing that sister chromatid cohesion is mediated by individual cohesin complexes. Read about it here: https://t.co/GDjy65YSn1 (1/4)
science.org
The sister chromatid cohesion necessary for faithful cell division is mediated by individual cohesin rings.
18
55
204
A reminder, we are currently searching for a #PhD and #postdoc to join us at the Center for Chromosome Stability in Copenhagen. The positions are fully funded. The deadline for the postdoc position is this Sunday (10.03.24). Details below:
Looking for a #postdoc or #PhD in eukaryotic #DNA replication and #CryoEM? We are recruiting! The positions are fully-funded by the @novonordiskfond and come with access to cutting-edge facilities and fantastic colleagues in #Copenhagen. See links below for more information.
0
11
22
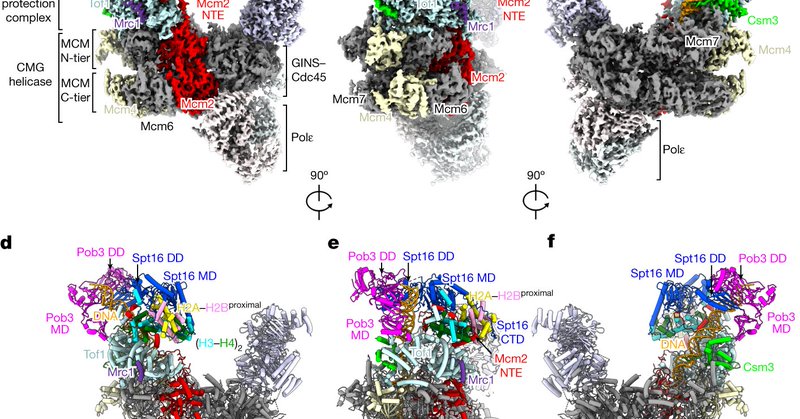
For those interested in chromatin, there are some incredible new papers in Nature describing decoding of chromatin states by chromatin readers ( https://t.co/VHHCKPV3Np) and parental histone transfer by the DNA replication machinery ( https://t.co/yWNnkVmbP8). Well worth a read!
nature.com
Nature - Structures of the yeast replisome associated with the FACT complex and an evicted histone hexamer offer insights into the mechanism of replication-coupled histone recycling for maintaining...
2
23
126
Fantastic opportunity! I would highly recommend working with Jan.
We are recruiting two PhD students 🎓 this round! Dry lab 💻: AI-based structural modeling method development and application. Wet lab 🧪: In-cell cryoET and/or crosslinking mass spectrometry of viral infection. Apply ✍️ by 11 March! ⌛️
0
1
5
Looking for a #postdoc or #PhD in eukaryotic #DNA replication and #CryoEM? We are recruiting! The positions are fully-funded by the @novonordiskfond and come with access to cutting-edge facilities and fantastic colleagues in #Copenhagen. See links below for more information.
5
50
73
A huge thanks to @chewtheng84 and all the members of the @NynkeDekkerLab, @CostaLabCrick and @DiffleyLab who have contributed to this project over the last few years. It has been a pleasure working with you all!
0
0
0
This explains asymmetric binding site affinities at yeast origins: CDK blocks re-replication (in part) by blocking MO, which is only required when one ORC site is low affinity. Thus, co-evolution of sequence-specific DNA binding by ORC and CDK regulation has shaped yeast origins.
1
0
0
We show that CDK phosphorylation of the Orc2 IDR inhibits MO formation and DH assembly on asymmetric origins (e.g. ARS1). MCM loading at origins with two high affinity sites is not blocked by CDK. Such origins are refractory to inhibition by CDK both in vitro and in vivo.
1
0
0
MO is not essential at origins with two high affinity sites, where two independently loaded MCM single hexamers (SHs) can assemble efficiently into an DH. We provide a high-resolution structure of the SH, which we argue is a key intermediate in all MCM loading pathways.
1
0
0