Stanislau Yatskevich
@StanislauY
Followers
221
Following
402
Media
12
Statuses
103
Post doc at Genentech, former PhD student at MRC-LMB
Joined August 2018
Exciting news - Our lab is recruiting a postdoc to join our explorations into the Plasmodium cytoskeleton. Do reach out if you'd like further details. Apply here; closing date 8th Sept: https://t.co/hyGSoTK9XG
2
10
25
We also discovered 4 previously uncharacterised proteins, which we named centromeric subunits 1-4 (CS module), functionally repurposed from the outer kinetochore Dam1/DASH complex. It will be very interesting to see how many other species contain this CS module
0
0
0
Interestingly, this species also lost centromeric CENP-A nucleosome, but instead evolved a unique histone fold protein that dimerises via non-canonical interfaces to fold the DNA
0
0
1
We discovered that holocentric kinetochore assembles into a self-contained, head-to-head dimer that topologically entraps and loops DNA, creating a point-centromere-like architecture.
2
0
2
Happy to share our latest work on the structure and assembly of holocentric kinetochores! Huge thanks to Ines for a very fruitful collaboration, Claudio for all the support, and congratulations to Christine and all co-authors! https://t.co/PP5oUXVdvW
1
6
36
Wonderful meeting at the Centromere Biology Gordon Conference in Maine! Lot of great science and nice friends to meet! Thanks @centromellone and @iaincheeseman for organizing it! @astraight @oegemadesai_lab @larsjansenlab @Foltz_Lab @StanislauY
1
1
38
New work from @palconh in my group shows that DNA repair proteins FANCD2-FANCI slide on double-stranded DNA. Surprisingly they specifically recognise single-stranded junctions #cryoEM Our first foray into single molecule imaging thanks to @RuedaLab @MRC_LMB @MRC_LMS Thread here:
Our latest work on FANCD2-FANCI (D2-I), a complex that helps resolve DNA crosslinks and replication stress, is out! https://t.co/bYoGGmbwkR We show D2-I acts as a sliding clamp on DNA, stalling at single-stranded gaps to identify and protect damage sites. @lapassmore @RuedaLab
6
18
110
Over and out for the Centromere Biology GRS! Great talks, great panel discussion, and a great intro to the GRC week ahead! Thanks to everyone who helped make these two days a success!
0
11
43
Last year, Stanislau Yatskevich (@StanislauY, then @MRC_LMB) won a Birnstiel Award for his groundbreaking research on chromosome segregation. His talk is now available online:
1
3
12
A gentle reminder that there is less than 1 month to register for the centromere biology GRS & GRC! The GRS is almost (!) fully subscribed so get your applications in ASAP if you wish to present a poster. We have a great lineup of speakers and mentors so don’t miss out!
Calling all aspiring centromere biologists! Rae Brown (Straight Lab) and I will co-chair the GRS for Centromere Biology, preceding the main GRC on 27th-28th July 2024. Please retweet and register before April 21st to be considered for a talk!
0
8
13
When I joined David Barford's group @MRC_LMB it was with the goal of using my cryo-ET and computational skills to help them understand funky biology. I joined @TDendooven and @StanislauY's efforts to average the native gamma tubulin ring complex (γTuRC) from tomograms of SPBs
1
25
163
Happy to see one of the last pieces of my PhD work published! Very lucky to have awesome @TDendooven as a partner in crime Also many thanks to @AlisterBurt, Dom, John, David and @RappsilberLab
Excited to see this work from @StanislauY and I out in @NatureSMB! We averaged the native yTuRC from Yeast SPBs, and discovered a novel coiled-coil protein stapling the Tubulin lattice to yTuRC. Huge thanks to @ABurt, David, John, @RappsilberLab and Dom! https://t.co/SpCaAtDdLt
1
5
22
Excited to see this work from @StanislauY and I out in @NatureSMB! We averaged the native yTuRC from Yeast SPBs, and discovered a novel coiled-coil protein stapling the Tubulin lattice to yTuRC. Huge thanks to @ABurt, David, John, @RappsilberLab and Dom! https://t.co/SpCaAtDdLt
6
29
99
Our new paper about the structural mechanism by which RAD51 assembles at double-stranded DNA break sites on chromatin has been published in Nature! https://t.co/bR8Oxjv8RP
#CryoEM #Nucleosome #RAD51 #DNArepair
11
117
482
Thank you! It was a pleasure to coordinate the publications
0
1
4
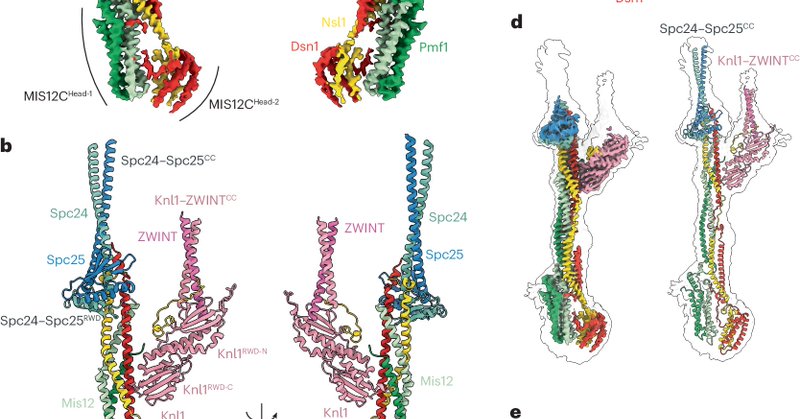
Happy to see our work on the structure of the outer kinetochore in print https://t.co/VvKmx2snJy Special thanks to David, Dom, @JYang188 and @ZhangZiguo Glad to see the complementary study by @AndreaMusacchi1,@polley_dilse and @TobiasRaisch in the same issue! Congratulations 🎉
nature.com
Nature Structural & Molecular Biology - Here, the authors determine the structure of the human outer kinetochore KMN network complex, showing that it forms an extended and rigid rod-like...
1
20
76
Congratulations @ElyseFischer7, @connyyu, David and co authors! Insightful work on the mechanism of Mad2 conversion by Cdc20 to a closed, active state 🎉
0
0
3
Very insightful work on the gamma-TuRC activation mechanism
Happy to share the first pre-print from the lab focusing on a closed conformation of the mammalian gamma-tubulin ring complex! https://t.co/ClZFneS1rJ
1
0
3
Phenomenal work on the yeast outer kinetochore! Congratulations 🎉
0
0
1