Sanchita Bhattacharya
@Sanchitab
Followers
105
Following
76
Media
1
Statuses
65
Join us for the invited talks from immunology experts and a hands-on session to explore open-access immunology data @immportdb using Advanced AI tools @chatgptapp @FOCISimmunology. Please register at https://t.co/phoW9iaTNN.
#AI #DataScience #FOCIS2024
0
0
2
🎉Exciting news! #ImmPortDB celebrates its 20th anniversary this year! 1k studies & vast amounts of immunologic & infectious disease data from 160+ areas & 35 types - is pivotal for global biomedical research! Visit: https://t.co/ji7cI6TH8z to learn more! #ImmPortDB #Research
niaid.nih.gov
This year, the immunology data repository ImmPort reaches a key milestone: 20 years since the first study was added to its repository in 2004. ImmPort is funded by NIAID’s Division of Allergy,...
0
3
6
CONGRATS @ImmPortDB (@NIAIDNews @PeratonCorp @UCSF_BCHSI) on reaching 1000 #openaccess #opendata sets related to immunology and inflammation and related conditions and treatments like cancer immunotherapy and pre-term birth! I still remember when we had fewer than 50! Some of
Huge congrats to #ImmPortDb data providers! ImmPort has now reached 1,000 studies because of your commitment to #FAIRdata sharing. Let's celebrate this remarkable achievement together! To learn more about ImmPort’s latest data release visit: https://t.co/KiUdkWi7pc
1
6
18
#ImmPortDB was thrilled to participate in DRKB- a meeting of NIH-affiliated repositories and knowledge bases. Thanks to @skleinstein for a great presentation on ImmPort during the Community-Driven Data Curation Panel, to the organizers, and to the @NIHDataScience for having us.
0
4
11
Thinking about applying for the Computational Models of Influenza Immunity (CMII) Cooperative Agreement Program at @NIAIDNews? See if @ImmPortDB #opendata on influenza and vaccination can help!
niaid.nih.gov
Propose a computational model that is data-driven, macromolecular, or hypothesis-based and mechanistically-driven and generalizable to multiple types of immune perturbations.
0
4
7
Finally available in California! Sign up to join the pilot California’s mobile driver’s license / ID card is a “free, secure, and convenient digital ID”, supposedly works at SFO and LAX. Doesn’t (yet?) work with Apple Wallet like other states…
dmv.ca.gov
The CA DMV Wallet app holds your mobile driver’s license (mDL) - a free, digital, secure, and convenient version of your California DL/ID.
3
4
17
Elegant #HumanImmuneVariation study by @lab_peters & @aidanbutty et al - mapping genetic determinants of plasma protein levels (@OlinkProteomics) https://t.co/0nvbRciLtA
@ImperialImmuno @imperialcollege
0
17
47
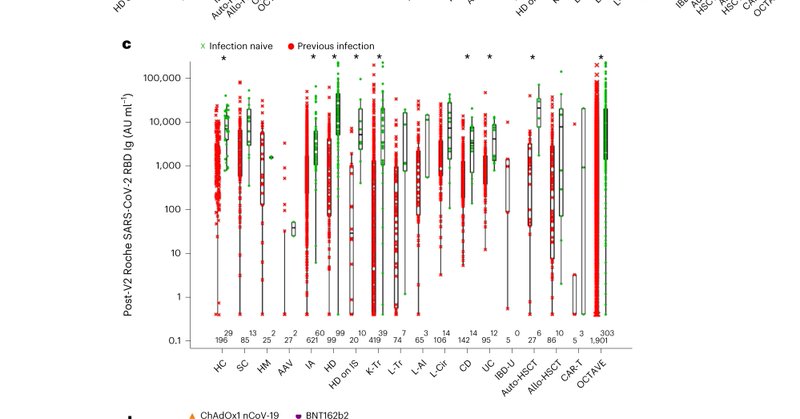
Delighted to share in Nature Medicine, COVID19 vaccine responses in >2800 immune suppressed people. Enormous thanks to patients and superb investigators @OUHospitals @OxfordBRC @SamMurray13 @tom_marjot @Georgi_Meacham @Susannajd and many more https://t.co/mGllALfCTA
nature.com
Nature Medicine - Serological analysis and infection outcomes of participants in the multi-center, prospectively enrolled OCTAVE cohort, comprising 2,686 participants with immune-suppressive...
4
34
110
Go see our partners #FOCIS2023 Build your skills and then tap our golden treasure of #immune data https://t.co/PC8bwABvpK
immunespace.org
Hello Boston! We are excited to see you tomorrow at #FOCIS2023 for #ImmPortDB's #BigData in #Immunology workshop. Join us to learn about public datasets, strengthen your funding requests, and build your analytical skills! There's still time! Register here https://t.co/BLswNYo40V
1
3
4
Legacy study Revealing #COVID19 Vaccine responses, broad/ crossprotection associated with specific B and T cell subsets, beautiful story 👇👇
🚨New preprint from Legacy study @TheCrick & @UCLHresearch. ⏩ https://t.co/tZkyFwmFes 👉We asked: "What's a 'normal' COVID vaccine response?" 🤔 Turns out there's 2 kinds of 'normal' in healthy adults 🧐 — and this has implications for protection vs. future variants 🤒.🧵 1/n
1
4
10
Actually, not transforming the data outperforms log(y/s+1). 1/
4
15
128
What a resource! 🤯 850,000 #scRNAseq cells from 226 samples across 10 cancer types!
New🔥 #DataScience #Bioinformatics resource: 850,000‼️ #scRNAseq cells from 226 samples across 10 cancer types draw a map of the tumor microenvironment, in particular fibroblasts. Let’s see👇what are the main contributions of this work & what this means for #cancer #Genomics🧵
0
4
8
If you want to explore the benchmark results yourself, check out our interactive shiny app at https://t.co/UmjBzjhfhL. All the code of the paper is available at https://t.co/X61ICd6sao.
0
3
13
Vaccine response #opendata in @ImmPortDB can even be used as a comparison group for new vaccine studies, as shown by Prabhu Arunachalam and team in @Nature: Systems vaccinology of the BNT162b2 mRNA vaccine in humans (paper https://t.co/dofJ8nBRWS)
#FOCIS2022
0
4
10
We just released #gget version 0.2.0! 🎉 Thanks to your feedback, we were able to further improve its ease of use. In addition to Ensembl IDs, gget now also supports WormBase 🪱 and FlyBase 🪰 IDs. All new features are listed here: https://t.co/9yFGH2gKiF
6
50
232
A big day for life science The Tabula Sapiens, like a Periodic Table of Human Cells, ~500,000 cells analyzed, 24 tissues / organs @ScienceMagazine "a broadly useful reference to deeply understand and explore human biology at cellular resolution" https://t.co/p0z103BnZH
16
353
1K
As we commemorate #InternationalWomensDay, we want to celebrate the wonderful women that lead and represent in #science and #research every day and make so many of the studies available in @ImmPortDB, possible. We thank you! #OpenScience #DataSharing #ImmPortDB
1
4
6
I still remember how this paper happened in lab meeting… congratulations @dvir_a and @ZichengHu!
Big milestone today 🔥🔥: the xCell paper just passed 1,000 citations on google scholar! 526 of them just this year. So great to see all the awesome papers using this tool we developed only 4 years ago. Stay tuned for xCell 2.0! @atulbutte @zichenghu
1
3
15
Really excited to share our paper on TetTCR-SeqHD! This new tech enables 4 dimensions of info on tens of thousands of single T cells, antigen specificity, TCR, gene exp, and phenotype. Thanks to all the authors, especially Keyue (Kevin) Ma for pushing it across the finish line!
Ma et al describe TetTCR-SeqHD - a method to simultaneously profile TCR sequences, cognate antigen specificities, targeted gene-expression, and surface-protein expression from tens of thousands of single cells. https://t.co/WqJafd86A1
9
35
148