Suresh Poovathingal
@S_Poovathingal
Followers
219
Following
315
Media
9
Statuses
274
Scientist, VIB Center for Brain and Disease Research
Louvain, Belgium
Joined May 2018
RT @shalinhnaik: A triumph of perseverance from twitterless Tom Weber, Christine Biben and the team, our in vivo barcoding "LoxCode mouse"….
0
54
0
RT @bettieliu: Delighted to share our latest work deciphering the landscape of chromatin accessibility and modeling the DNA sequence syntax….
biorxiv.org
Transcription factors (TFs) establish cell identity during development by binding regulatory DNA in a sequence-specific manner, often promoting local chromatin accessibility, and regulating gene...
0
65
0
RT @satijalab: Interested in single cell genomics but need help getting started? Check out the full agenda for our Single Cell Genomics Day….
0
206
0
RT @satijalab: Interested in single cell genomics but need help getting started? Check out my lab's Single Cell Genomics Day on April 25. T….
0
241
0
RT @naturemethods: The wait is over!! We are thrilled to announce that we have chosen Spatial Proteomics as 2024’s Method of the Year! 🥳. F….
0
300
0
RT @LGMartelotto: 💥🚀. Sub-cellular Imaging of the Entire Protein-Coding Human Transcriptome (#18933-plex) on FFPE Tissue Using Spatial Mole….
biorxiv.org
Single-cell RNA-seq revolutionized single-cell biology, by providing a complete whole transcriptome view of individual cells. Regrettably, this was accomplished only for individual, tissue-dissocia...
0
10
0
RT @LGMartelotto: Sainsc: A Computational Tool for Segmentation‐Free Analysis of In Situ Capture Data - Müller‐Bötticher - Small Methods -….
onlinelibrary.wiley.com
Spatially resolved transcriptomics methods can now profile the entire transcriptome, at full organism scale with nanometre resolution. Analysis frameworks that can efficiently and intuitively...
0
5
0
RT @fabian_theis: Need to design your next Xenium/CosMx/Merfish experiment? Spapros, now out @naturemethods, allows for probe set selection….
0
71
0
Happy to see our work, Nova-ST, a new open-source platform for high-resolution spatial transcriptomics published in @CellCellPress @CellRepMethods . 👇.
cell.com
Poovathingal et al. develop Nova-ST, a low-cost, whole-transcriptome spatial workflow based on Illumina NovaSeq flow cells. The nano-patterned structure allows high-resolution, single-cell spatial...
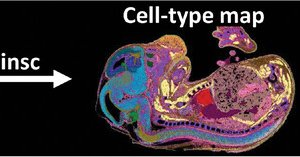
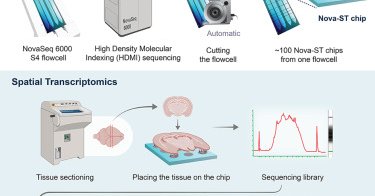
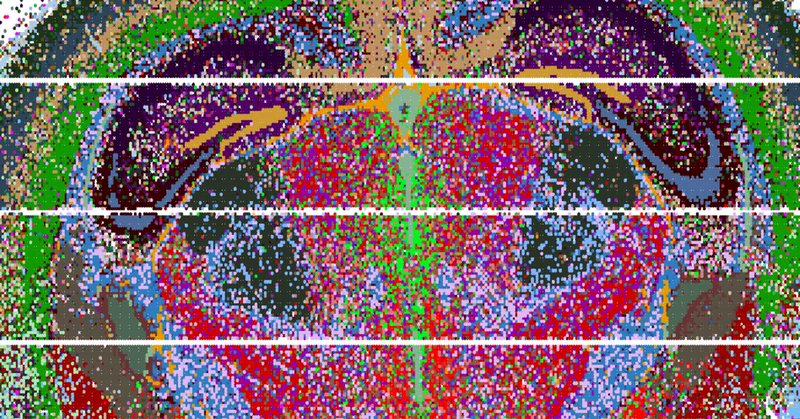
🗺 Introducing Nova-ST, a new open-source platform for high-resolution spatial transcriptomics! ⚡ Developed in the @steinaerts lab, Nova-ST will make large-scale, high-resolution spatial tissue analysis more accessible and affordable. Find out more ➡️
6
12
52
RT @LGMartelotto: Accurate quantification of single-cell and single-nucleus RNA-seq transcripts using distinguishing flanking k-mers | bioR….
biorxiv.org
In single-cell and single-nucleus RNA sequencing, the coexistence of nascent (unprocessed) and mature (processed) mRNA poses challenges in accurate read mapping and the interpretation of count...
0
8
0
RT @CalebLareau: On bioRxiv today, we raise serious concerns over a recent @nature paper describing ReDeeM, a new single-cell mitochondrial….
biorxiv.org
Sequencing mitochondrial DNA (mtDNA) variants from single cells has resolved clonality and lineage in native human samples and clinical specimens. Prior work established that heteroplasmic mtDNA...
0
118
0
RT @fabian_theis: Happy that CellRank2 is out @naturemethods! @PhilippWeiler7 & @MariusLange8 led this multimodal extension of CellRank, to….
0
59
0
RT @GenomeWeb: Researchers Turn Illumina's NovaSeq 6000 Into Spatial Transcriptomics Instrument
genomeweb.com
For intrepid scientists, the hardware hack may unlock larger spatial biology studies that would be too expensive to run on commercial spatial omics platforms.
0
3
0
RT @biorxiv_genomic: Droplet Hi-C for Fast and Scalable Profiling of Chromatin Architecture in Single Cells #biorx….
0
2
0
10/ More details on Nova-ST Data analysis and processing:.
github.com
A repository containing the analysis scripts for Nova-ST data - aertslab/Nova-ST
0
0
1
9/ Detailed protocols can be found here:.Nova-ST chip preparation:.Nova-ST spatial transcriptomics workflow:.Keep an eye out for the future developments we are currently performing to make the Nova-ST workflow much more simpler !!.
protocols.io
Nova-ST is a an open-source, high-resolution sequencing based spatial transcriptomics workflow. This method gives comparable resolution to BGI Stereoseq, SeqScope & PIXEL se...
1
0
1