Roy Ξ
@Roy_ehling
Followers
118
Following
186
Media
8
Statuses
119
Protein Engineering - Immunotechnology. Small biotech (He/Him). Opinions are my own.
Germany, Switzerland
Joined May 2011
New research on Senescent-resistant MSCs (SRC) in Cyno monkeys: treating age-related illness. Huge study by Lei et al @altos_labs #aging #dontdie
https://t.co/eFAfZkRAqC Could MSC/SRC exosome therapy become a thing for systemic rejuvenation? @bryan_johnson
0
0
0
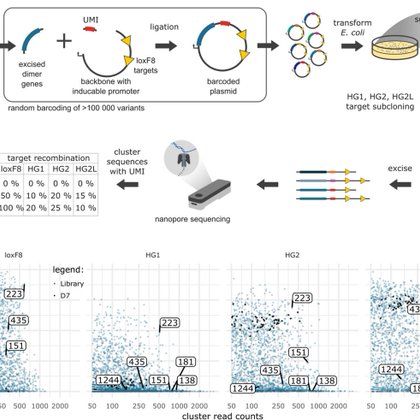
Excited to share my first last Author paper. Out now in @GenomeBiology We @BuchholzLab_TUD report two new methods. To evolve CRISPR-Cas systems (CaSLiDE) and to screen thousands of variants at once (DEQSeq) Thanks to the amazing team of co-authors!
genomebiology.biomedcentral.com
We introduce DEQSeq, a nanopore sequencing approach that rationalizes the selection of favorable genome editing enzymes from directed molecular evolution experiments. With the ability to capture...
1
3
15
My dad, who is a moderate Muslim, finally expressed his concern and asked me, 'Why are you supporting this genocide?' He mentioned that he's stopped looking at his phone because he can no longer bear to watch videos of Palestinian children being bombed by Israel. I replied, 'I
4K
25K
81K
The Flotilla 13 elite unit was deployed to the area surrounding the Gaza security fence in a joint effort to regain control of the Sufa military post on October 7th. The soldiers rescued around 250 hostages alive. 60+ Hamas terrorists were neutralized and 26 were
3K
11K
57K
Here's the preprint by @BenjMurrell and colleagues now posted, summarized in his thread https://t.co/6kuQCL8AJs "BA.2.75.2 was neutralised...five-fold less potently than BA.5... These data raise concerns that BA.2.75.2 may effectively evade humoral immunity in the population."
biorxiv.org
Several sublineages of omicron have emerged with additional mutations that may afford further antibody evasion. Here, we characterise the sensitivity of emerging omicron sublineages BA.2.75.2,...
4
37
107
ETH-Zürich (@ETH_en) has put out a great summary of our DML technique and how it can help us develop more effective antibody therapies and vaccines against future variants of SARS-CoV-2. Read more about it here:
0
2
1
Fantastic new tool developed by the team of Sai Reddy @ReddyLab_ETHZ: "#MachineLearning could support #antibody #drug development by enabling researchers to identify which antibodies have the potential to be most effective against current and future #Variant|s" @saireddy911
Researchers have developed a method to explore the possibilities of how the #pandemic #virus could evolve. Thanks to their work, it may be possible to develop #antibody #therapies and #vaccines that are more effective also against future variants. https://t.co/QxWQgM4FV0
0
5
11
Happy to share our recent manuscript from @ReddyLab_ETHZ has been published in Cell! Great work by @JosephMTaft & @cedricrweber et al. Moving towards predicting immune escape of future viral variants🤓
Now online! Deep Mutational Learning Predicts ACE2 Binding and Antibody Escape to Combinatorial Mutations in the SARS-CoV-2 Receptor Binding Domain
0
4
17
Sone good news about the BA.2.75 variant, in case it gets legs: it's not more immune evasive than BA.5, has cross-immunity w/ BA.1/2, and is still sensitive to Evusheld and Bebtelovimab https://t.co/EgnyuWpCYm
@TheLancetInfDis
@BenjMurrell @PeacockFlu @DannySheward & colleagues
40
344
1K
Update: Our deep mutational learning #DML paper on #SARSCoV2 profiling is now online at @CellCellPress! Now with more in-depth assessments of a greater number of antibodies! https://t.co/SIh8ltLl2E
cell.com
A machine-learning-guided, protein engineering method enables the prediction of how SARS-CoV-2 RBD combinatorial mutations will impact therapeutic antibody escape and ACE2 affinity. This method...
1
9
27
Proud to share our *finally* accepted paper. It was a wild ride, but we finally got there on my last day at @ETH_BSSE @ETH. Couldn't ask for a better way to finish my time in the @ReddyLab_ETHZ. https://t.co/eAz9AHZLq4
cell.com
A machine-learning-guided, protein engineering method enables the prediction of how SARS-CoV-2 RBD combinatorial mutations will impact therapeutic antibody escape and ACE2 affinity. This method...
1
11
51
Evasion of neutralizing antibodies by Omicron sublineage BA.2.75
biorxiv.org
Towards the end of 2021, SARS-CoV-2 vaccine effectiveness was threatened by the emergence of the Omicron clade (B.1.1.529), with more than 30 mutations in spike. Recently, several sublineages of...
0
31
54
Very happy that our work on generative machine learning for antibody design is now out in mAbs https://t.co/mCpEInbBZa. All details are in the preprint thread below. The peer-reviewed version contains more controls and also more comparisons of synthetic and experimental data.
tandfonline.com
Generative machine learning (ML) has been postulated to become a major driver in the computational design of antigen-specific monoclonal antibodies (mAb). However, efforts to confirm this hypothesi...
3
42
120
Protein engineers work wonders with machine learning, but generating data remains a bottleneck. Computer vision & NLP use data augmentation, but we lack such techniques for proteins. To address this, we develop nucleotide augmentation (NTA). A thread 🧵1/5
biorxiv.org
Machine learning-guided protein engineering is a rapidly advancing field. Despite major experimental and computational advances however, collecting protein genotype (sequence) and phenotype (functi...
1
8
25
Platypus intensifies..
Some work lead by fellow platypus enthusiast @Roy_ehling from @ReddyLab_ETHZ. We profiled repertoire & functional properties of PCs in convalescent COVID19 patients Adds to our previous characterisation of VDJ & GEX of T and B cells in @flobdalabob et al https://t.co/ylttnVIET1
0
0
3
SARS-CoV-2 reactive and neutralizing antibodies discovered by single-cell sequencing of plasma cells and mammalian display
0
6
11
Highlighting our recent work from @ReddyLab_ETHZ involving the construction of combinatorial RBD libraries that were used to infer and validate ACE2 binding and escape from clinically relevant mAbs. Moving towards in silico prediction of escape for emerging variants🤓
Predictive profiling of SARS-CoV-2 variants by deep mutational learning https://t.co/NNAeZFDfrT
#bioRxiv
0
6
24
@JosephMTaft @ReddyLab_ETHZ We also found structural evidence that higher-order network effects between the Omicron mutated sites may perturb the physical and chemical properties of the epitope surface for this mAb. https://t.co/xfn3r07buh
0
1
4
Why Deep Mutational Learning matters, a story in two press releases: https://t.co/9ePpxdRGwo
biorxiv.org
The continual evolution of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and the emergence of variants that show resistance to vaccines and neutralizing antibodies ( [1][1]–[4][2]...
0
4
9