Sai Reddy
@saireddy911
Followers
830
Following
280
Media
8
Statuses
91
Associate Professor - Swiss Federal Institute of Technology Zurich @ETH_en
Basel, Switzerland
Joined March 2016
Predicting TCR antigen specificity at proteome‑scale with synthetic immune cells and machine learning. I’m pleased to share our group's collaborative effort with Engimmune in this new pre‑print:
biorxiv.org
TCR specificity to peptide-HLA antigens is central to immunology, impacting responses in infection, autoimmunity and cancer. Achieving precise recognition while avoiding off-target reactivity is...
0
2
10
Sharing our computational immuno ecosystem - started for internal use but might help others too! CRAN package >100 functions (both custom&external tools eg cellphoneDB,projecTILs,alphafold) Database to download&integrate scSeq/VDJ directly into R + ~20 tutorials w/ code,data,text
ePlatypus: an ecosystem for computational analysis of immunogenomics data https://t.co/VYM9sd6V92
#biorxiv_immuno
4
32
119
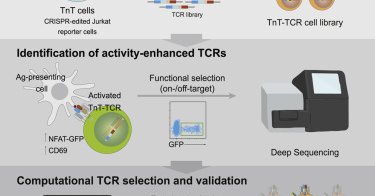
Online now: High-throughput T cell receptor engineering by functional screening identifies candidates with enhanced potency and specificity
cell.com
Vazquez-Lombardi et al. developed a high-throughput method combining mammalian display, CRISPR-targeted mutagenesis, functional screening, and deep sequencing for the engineering of T cell receptors...
0
20
64
Update: Our deep mutational learning #DML paper on #SARSCoV2 profiling is now online at @CellCellPress! Now with more in-depth assessments of a greater number of antibodies! https://t.co/SIh8ltLl2E
cell.com
A machine-learning-guided, protein engineering method enables the prediction of how SARS-CoV-2 RBD combinatorial mutations will impact therapeutic antibody escape and ACE2 affinity. This method...
1
10
27
Proud to share our *finally* accepted paper. It was a wild ride, but we finally got there on my last day at @ETH_BSSE @ETH. Couldn't ask for a better way to finish my time in the @ReddyLab_ETHZ. https://t.co/eAz9AHZLq4
cell.com
A machine-learning-guided, protein engineering method enables the prediction of how SARS-CoV-2 RBD combinatorial mutations will impact therapeutic antibody escape and ACE2 affinity. This method...
1
11
51
Protein engineers work wonders with machine learning, but generating data remains a bottleneck. Computer vision & NLP use data augmentation, but we lack such techniques for proteins. To address this, we develop nucleotide augmentation (NTA). A thread 🧵1/5
biorxiv.org
Machine learning-guided protein engineering is a rapidly advancing field. Despite major experimental and computational advances however, collecting protein genotype (sequence) and phenotype (functi...
1
8
25
Landing therapeutic genes safely in the human genome https://t.co/8QOkbDeKnF via @wyssinstitute
wyss.harvard.edu
By Benjamin Boettner (BOSTON) — Many future gene and cell therapies to treat diseases like cancer, rare genetic and other conditions could be enhanced in their efficacy, persistence, and predictabi...
0
0
4
Highlighting our recent work from @ReddyLab_ETHZ involving the construction of combinatorial RBD libraries that were used to infer and validate ACE2 binding and escape from clinically relevant mAbs. Moving towards in silico prediction of escape for emerging variants🤓
Predictive profiling of SARS-CoV-2 variants by deep mutational learning https://t.co/NNAeZFDfrT
#bioRxiv
0
6
25
The lesson: individual mutations cannot predict the behavior of combinatorial mutations. Read more: https://t.co/S8DQPLLgib
https://t.co/9ePpxdRGwo
@ReddyLab_ETHZ
biorxiv.org
The continual evolution of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and the emergence of variants that show resistance to vaccines and neutralizing antibodies ( [1][1]–[4][2]...
1
3
7
Spinoff deepCDR Biologics acquired by Boston-based company Alloy Therapeutics @AlloyTx The acquisition of #deepCDR adds expertise in #Bioinformatics and #MachineLearning to Alloy’s #antibody discovery workflow. > https://t.co/WuIdj0cEIy
#ETHspinoff @ETH_en @ReddyLab_ETHZ
0
6
15
Another Omicron neutralization thread, this time about monoclonal antibodies. Once again, massive cred to @DannySheward, and everyone else from the previous data release. This time, we also need to especially thank @ReddyLab_ETHZ (1/n)
1
8
17
As reported in the Economist this summer, it may indeed be possible to predict viral evolution by combining #AI and #ML methods. Check out our latest preprint: https://t.co/eKOh4ANzA1
biorxiv.org
The continual evolution of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and the emergence of variants that show resistance to vaccines and neutralizing antibodies ( [1][1]–[4][2]...
0
0
5
Predictive profiling of SARS-CoV-2 variants by deep mutational learning https://t.co/NNAeZFDfrT
#bioRxiv
biorxiv.org
The continual evolution of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and the emergence of variants that show resistance to vaccines and neutralizing antibodies ( [1][1]–[4][2]...
1
3
14
We are thrilled to join Alloy on its mission of empowering scientific entrepreneurs and democratizing foundational drug discovery capabilities
businesswire.com
Alloy Therapeutics, a biotechnology ecosystem company, announced it has acquired deepCDR Biologics, a Basel, Switzerland-based developer of deep learning tec...
1
2
20
Catch Prof. @saireddy911's and Dr. @JosephMTaft's presentations at #PEGS Europe 2021 in Barcelona. When: today Topic: Predictive profiling of #SARS evolution using #DeepLearning Find us during the poster sessions this afternoon (posters 11,12,13,14). See you there!
0
4
6
DeepSARS: new tool developed with @TanjaStadler_CH, @LabOxenius, F. Rudolf, @AlexYermanos and several other great collaborators, we hope to improve #COVID19 diagnostics & early detection of #variants by genomic surveillance of #coronavirus #SARSCoV2
https://t.co/U5nvf2QDwt
2
15
33
🤔 Combating future viruses – Predicting viral evolution may let vaccines be prepared in advance
economist.com
New techniques could programme people’s immune systems against future pathogens
9
1
9