𝓟𝓮𝓷𝓰 𝓗𝓮 何鵬
@PengHeAtlas
Followers
1K
Following
1K
Media
119
Statuses
1K
bioinformatics for multiome➕spatial➕single-cell; development↔disease; Assistant professor at UCSF; Tweets👉own; BlueSky: https://t.co/5yOclEVU7u Mastodon:penghe
San Francisco, CA
Joined January 2013
RT @KexinHuang5: 🧬 Excited to open-source Biomni! With just a few lines of code, you can now automate biomedical research with AI agent!. W….
0
98
0
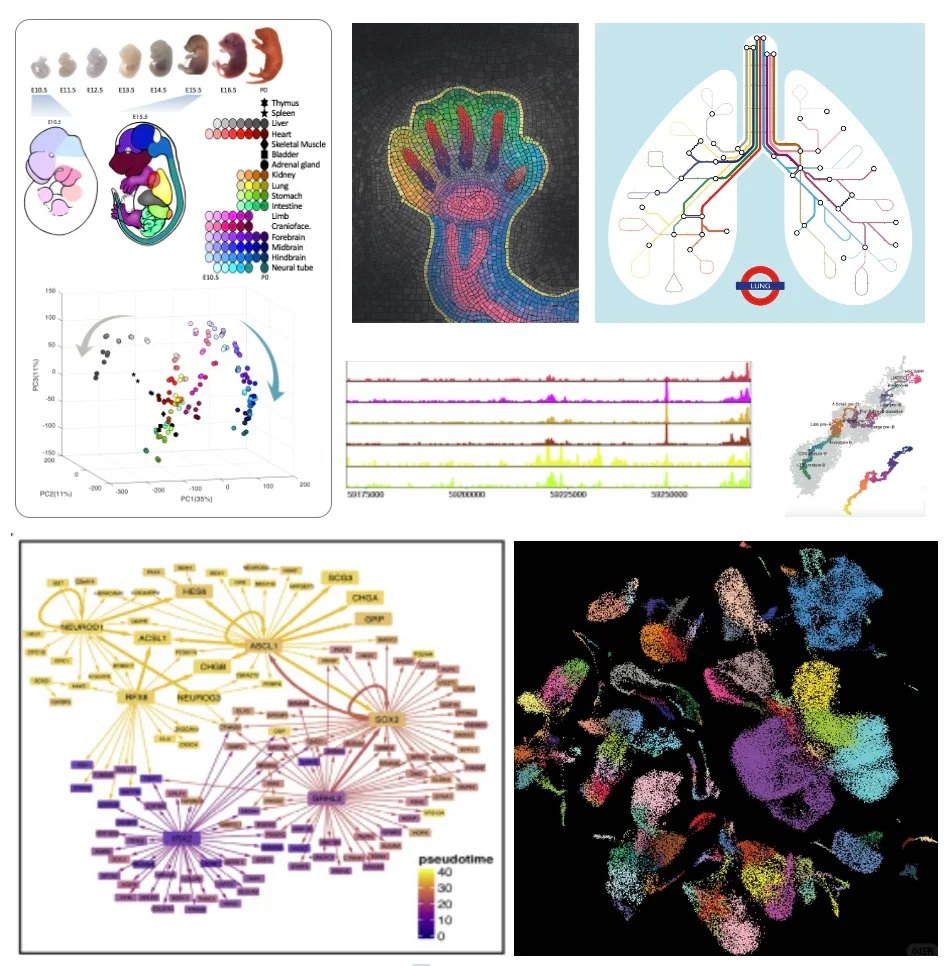
Great paper proposing a graph/neighborhood-based way to supplement current metrics in single-cell integration benchmarking. Excited to see our fetal lung cell atlas used to guide metric evaluation. Biology is continuous—and so should be our thinking, including in AI. 👏🧬🤖.
Defining “what is good” is central to both AI and Science. Despite massive efforts towards building the Virtual Cell, our recent @NatureBiotech article reflects on whether current gold standards (i.e., evaluation metrics) still hold up.
0
0
14
RT @KexinHuang5: 📢 Introducing Biomni - the first general-purpose biomedical AI agent. Biomni is built on the first unified environment fo….
0
115
0
RT @SikkemaLisa: 1/7 Planning to build a single-cell atlas? Or wondering how atlases can be useful to your research? Read our guide on sing….
0
17
0
Great opportunity!.
We are hiring! Join us in building virtual embryo models — an ambitious and exciting mission that goes beyond the widely acclaimed “virtual cell” initiative! 🚨. Are you a technology developer, developmental biologist, or machine learning expert interested in creating predictive,
0
0
2
RT @s_h_saunders: Please help me amplify a special faculty search. We're looking for someone in "Microbial Systems Biology" to join me and….
0
39
0
RT @MartinUCSF: Are you interested in "skipping the postdoc" and starting your independent lab in biomedical research as a fellow?. Check o….
0
113
0
And kudos to the co-drivers of the project David Horsfall,.@Muzz_Haniffa and Daniela Basurto-Lozada.for finishing the paper!.
0
0
2
PS please check out a complementary visualization tool here:
Using the 🆕 spatial transcriptomics feature in CZ #CellxGene, we reproduced the key finding of a study from @PengHeAtlas in the Teichmann lab showing transcriptionally + spatially distinct mesenchyme populations in the autopod. Explore the dataset ➡️
0
0
2
PS. Please also check out another amazing tool here:
Happy to see the final version online! Great visualization tool developed by @bayraktar_lab and led by @BioinfoTongLI #team #Science.
0
0
0
Can't emphasize enough that this is a great platform. Really appreciate the effort of everyone (CZI Cellxgene team and @martinprete for developing the features, as well as my student Weimin Lin and HCA Lattice for formatting the data).
Using the 🆕 spatial transcriptomics feature in CZ #CellxGene, we reproduced the key finding of a study from @PengHeAtlas in the Teichmann lab showing transcriptionally + spatially distinct mesenchyme populations in the autopod. Explore the dataset ➡️
2
0
5
Happy to see the final version online! Great visualization tool developed by @bayraktar_lab and led by @BioinfoTongLI #team #Science.
2
5
28
Very happy to have contributed to showcasing the 🆕 features of this incredible platform 🌟, dedicated to making basic single-cell/nucleus and🗺️spatial transcriptomics analysis accessible to everyone, regardless of programming skills 💻!!.
We were able to quickly reproduce key findings from @PengHeAtlas of the @teichlab’s new work on human limb development using CZ #CellxGene’s new spatial features. Give it a try and, as always, please let us know what you think
0
1
19
🚨𝐇𝐞 𝐋𝐚𝐛 job opportunity @UCSF 🚨:.I'm hiring TWO postdocs - an experimentalist and a computational biologist. Join us to use cutting-edge single-cell and spatial technologies to unravel the mysteries of human tissues.
1
92
199
RT @CDominguezConde: 📢#PhD position available in the CDC lab focused on modelling the effect of genetic variation on T cell development and….
0
25
0