Can Ergen
@_canergen
Followers
215
Following
95
Media
4
Statuses
90
Postdoc at @UCBerkeley and @UCSF. Co-mentored by @Yoseflab and @yimmieg
Berkeley
Joined November 2018
RT @stats_stephen: Important topic, but this is more of a quick-start guide. For cutting-edge research on LLM evals, see these papers usin….
0
6
0
RT @dianyang1117: Excited to share our work on a new spatial-lineage platform for tumor evolution analysis led by very talented postdocs @M….
0
5
0
RT @scverse_team: Big thanks to @_lazappi_ for being part of the first-ever #scverse2024 conference and for the amazing summary and sketch….
lazappi.id.au
Recap of the first scverse conference and sketchnotes of keynote talks
0
3
0
RT @scverse_team: The first scverse community survey is LIVE!📋🐍🧑💻 Do you use or contribute to scverse tools for omics analysis? Your feedb….
docs.google.com
In an effort to better understand the scverse community, we (the scverse core team) have created this short survey. It should take you less than 5 minutes to complete. We will use the results from...
0
8
0
RT @MKoutrouli: First conference, first selfie with some of the incredible @scverse_team core and volunteers! Feeling so grateful to be par….
0
7
0
RT @NeBanovich: Earlier this week we carried out a preliminary analysis of our first @10xGenomics Xenium prime (5,000 gene) run. We had des….
0
34
0
RT @LukasHeumos: I'm stoked to announce our new end-to-end framework for perturbation analysis! Our scverse advisory committee highlighted….
0
26
0
RT @CalebLareau: Somatic mutation tracing with single-cell data is a fairly niche but growing field. We can learn more from each other via….
0
4
0
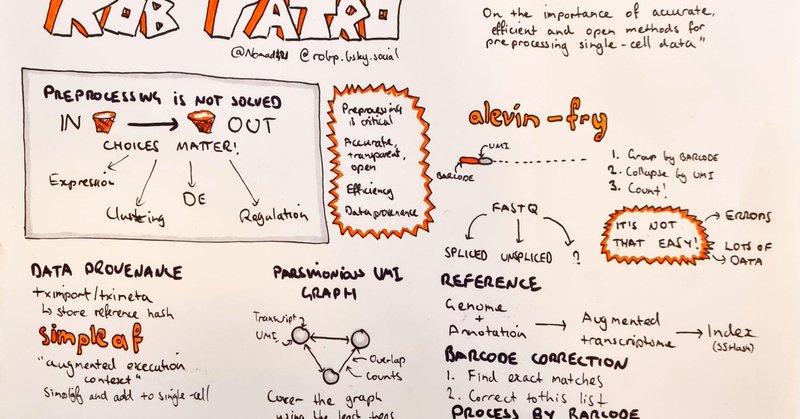
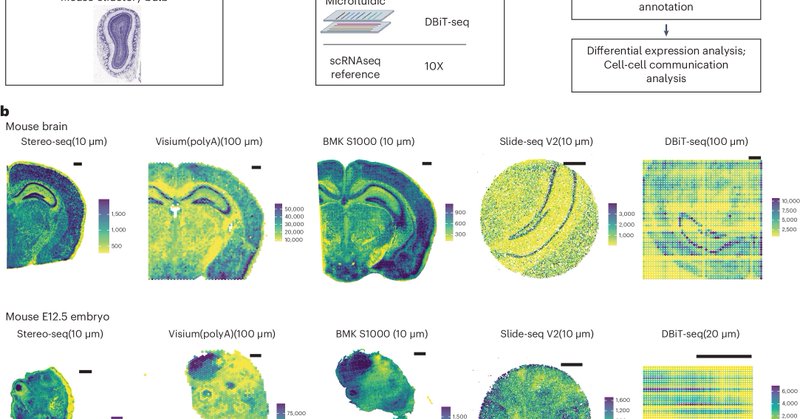
RT @LGMartelotto: Systematic comparison of sequencing-based spatial transcriptomic methods | Nature Methods
nature.com
Nature Methods - This analysis presents a systematic comparison of 11 sequencing-based spatial transcriptomics methods using well-characterized references, which offers insights into performance...
0
27
0
RT @nathan_levyy: So proud of my intern @Rotem_Shalita for her work at the #AiHub on deciphering T cell homing signatures for optimized CAR….
0
2
0
RT @Severin_1991: 🌟 Join me at #Scverse2024 for the "GPU-Accelerated Single-Cell Analysis with rapids-singlecell" workshop! .📍 Munich, Germ….
github.com
rapids_singlecell: GPU-accelerated framework for scRNA analysis - scverse/rapids_singlecell
0
11
0
RT @N_Rajewsky: i am so excited about our open source inexpensive, sub-cellular capture, do-it-yourself Open-ST that transforms tissues int….
0
93
0
RT @LukasHeumos: Our @scverse_team zulip chat recently crossed 500 members. It's super cool to have so many developers and users in one pla….
scverse.zulipchat.com
Browse the publicly accessible channels in scverse without logging in.
0
4
0
RT @MKoutrouli: The end of an era! The PhD is now gone but everything I’ve learned and the people I’ve met will stay! What a nice reflectio….
0
8
0
RT @xarray_dev: We are looking for an Xarray community developer to work at @EarthmoverHQ to focus on building connections, both technical….
0
19
0
RT @NicolaMTomas: Truly happy to see this out @NEJM - translational work from @Tobias_B_Huber and my lab on autoantibodies in nephrotic syn….
0
6
0
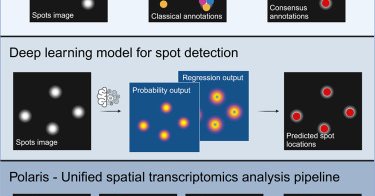
RT @davidvanvalen: I'm super happy to see part of our lab's work out this week in Cell Systems. Emily Laubscher's work presents a compellin….
cell.com
Polaris is a deep-learning-based analysis pipeline for spatial transcriptomics images. Polaris integrates a weakly supervised deep-learning model for spot detection with cell segmentation and gene...
0
12
0
RT @scverse_team: @Severin_1991, @ivirshup, Ilan Gold and @felix_f0097 teamed up at the #OpenHackathons in Görlitz, hosted by @casusscienc….
0
4
0
RT @mo_lotfollahi: Today, May 12, is Mother’s Day. A special mother and brilliant mathematician was also born today: Maryam Mirzakhani, the….
0
31
0
RT @AshleyKiemen: I’m beyond excited to share work from my PhD, co-led with Alicia Braxton, out today in Nature! We used CODA to map the an….
0
50
0