Kasit Chatsirisupachai
@KasitC10
Followers
331
Following
3K
Media
16
Statuses
308
Postdoctoral Fellow at the Genome Biology Unit @EMBL Heidelberg. Slowly migrating to https://t.co/khL5DLYjGK
Heidelberg, Germany
Joined August 2017
Excited to see this out in print @MolSystBiol! A great team effort especially with @ChristineMoene @molina_lab Arnaud. @EMBLHeidelberg.
Excited to share the latest story from the @arnaud_kr lab @embl ! Together with @ChristineMoene we used Single Molecule Footprinting to reveal that RNA Pol II occupancy occurs at surprisingly low-frequency at mammalian promoters! Thread below (⬇️) 1/11
1
2
20
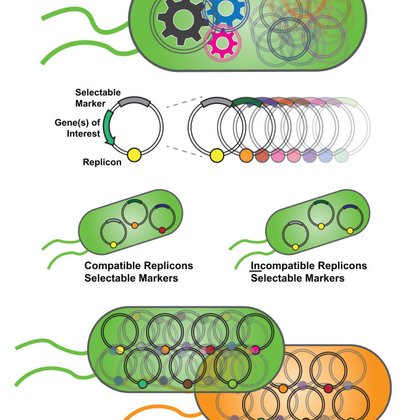
RT @O_Borkowski: How many plasmids can bacteria carry? A synthetic biology perspective.
royalsocietypublishing.org
Plasmids are pinnacle tools in synthetic biology and other biotechnological applications. They serve as the simplest approach to introduce recombinant DNA, which is then transcribed into RNA that...
0
66
0
RT @STOPlabPI: 🚨New preprint from our labs (STOP lab & Nojima lab @pol2rna)! @kopczynska_m and Chihiro Nakayama have been digging into how….
0
14
0
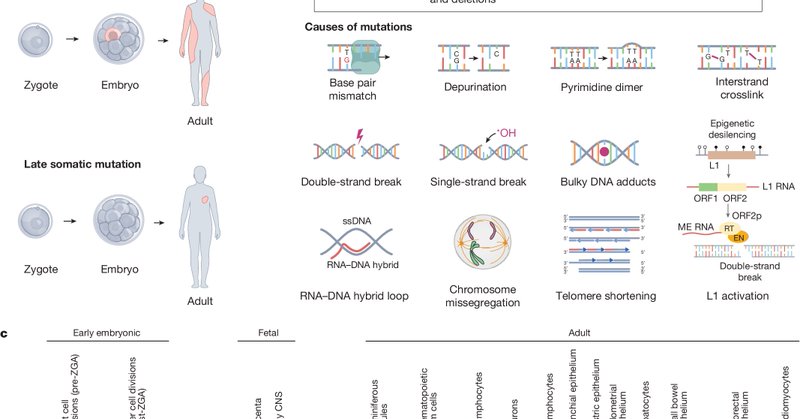
RT @TimCoorens: The cells in our bodies constantly acquire mutations. But what are the patterns of mutations across tissues? How do mutatio….
nature.com
Nature - The Somatic Mosaicism across Human Tissues Network aims to create a reference catalogue of somatic mosaicism across different tissues and cells within individuals.
0
41
0
RT @FukayaLab: Delighted to share a new review paper from the lab. We discussed recent key findings in enhancer biology. Thanks Dan Larson….
0
14
0
RT @BarzaghiGuido: Having a stressful day? Lay back, crack a beer open and read about how chromatin accessibility responds to TF binding p….
0
10
0
RT @JonasKoeppel: I’m delighted to share our work on scrambling the human genome using prime editing, repetitive elements, and recombinases….
0
83
0
RT @zanehkoch: WHY DOES THE EPIGENETIC CLOCK TICK?. While money has been poured into developing therapies to reverse the epigenetic clock (….
0
213
0
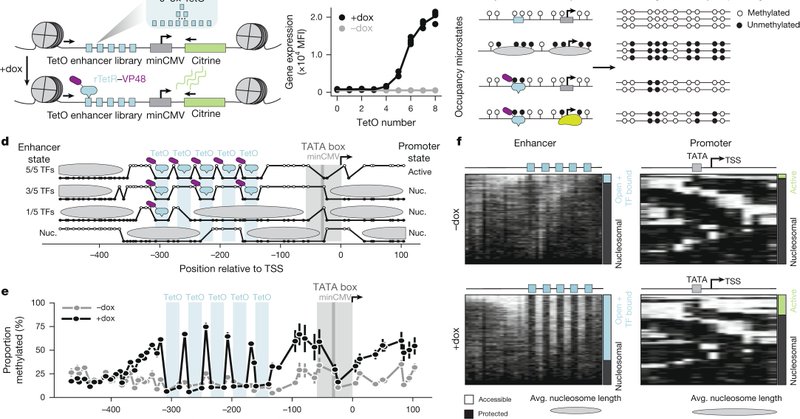
RT @WJGreenleaf: Incredibly excited that our paper linking single molecule states of TF binding to gene expression using quantitative therm….
nature.com
Nature - A study uses single-molecule footprinting to measure protein occupancy at regulatory elements on individual molecules in human cells and describes how different properties of transcription...
0
108
0
Finally I met @tangming2005 or “crazyhottommy” in person, a real pleasure! When I first start doing genomics/bioinformatics, I learnt a lot from your blog posts, tutorials, and github. Thanks a lot!
2
1
21
RT @TimCoorens: Very happy and excited to announce that I'll be starting my own research group at @emblebi! The group will focus on lineage….
0
31
0
RT @EMBO: More than 100 @EMBOFellows are meeting in Heidelberg. The postdoctoral fellows present their research, attend a mentoring session….
0
26
0
RT @ItaiYanai: Doing good science is 90% finding a science buddy to constantly talk to about the project.
0
3K
0
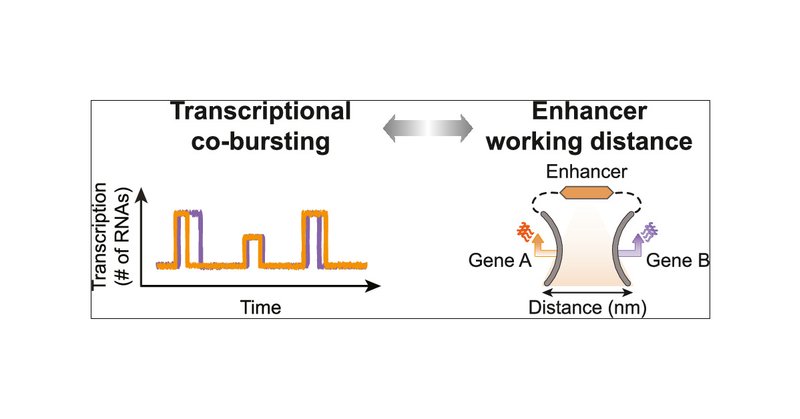
RT @cbohrer3141: I've always been obsessed with how genes and enhancers talk to each other. Our latest review explores just that. Enhancer….
pubs.acs.org
Enhancers are central for the regulation of metazoan transcription but have proven difficult to study, primarily due to a myriad of interdependent variables shaping their activity. Consequently,...
0
49
0
RT @ChristineMoene: Did you ever wonder how transcription initiation compares between flies and mammals? The project that started as my mas….
biorxiv.org
The general transcription machinery and its occupancy at promoters are highly conserved across metazoans. This contrasts with the kinetics of mRNA production that considerably differ between model...
0
4
0
RT @SchubelerLab: Great to see that the Krebs lab continues to provide quantitative and important insights into rate-limiting steps in tran….
0
2
0
@arnaud_kr @embl @ChristineMoene @molina_lab @ElisaKre Finally, I’ll be presenting this project at the EMBL Quantitative biology to molecular mechanisms conference in November. Please stop by if you are around!. 11/11.
embl.org
2
0
6
@arnaud_kr @embl @ChristineMoene @molina_lab It’s been an incredible journey working on this project! Huge thanks to the co-authors Christine, @ElisaKre , Roos, Nacho, and Arnaud. Please check out our preprint at 10/11.
biorxiv.org
The general transcription machinery and its occupancy at promoters are highly conserved across metazoans. This contrasts with the kinetics of mRNA production that considerably differ between model...
1
1
13
@arnaud_kr @embl @ChristineMoene @molina_lab Incorporating the turnover rate into our model suggested that it cannot fully explain the difference in Pol II occupancy. The remaining variance likely comes from a lower transcription initiation rate in mouse. 9/11
1
0
3