Hyunkyung Kim
@Hyunkyungkim_
Followers
121
Following
111
Media
1
Statuses
23
Genetics PhD Student in @AndyWDahl lab @UChicagoGGSB 🧬👩🏻💻
Chicago, IL
Joined December 2020
When you have a great software / method name: #SPATULA but next need to work out what it means hehe 😅 @Hyunkyungkim_ made us all laugh 🤣 #BoG24
1
3
18
Exciting to see this in print! Lots of gratitude for the amazing teamwork that made this possible. If you are looking to apply the #multiancestry #T2D genetic process-specific #polygenicscores to your work, variant weights are also available here:
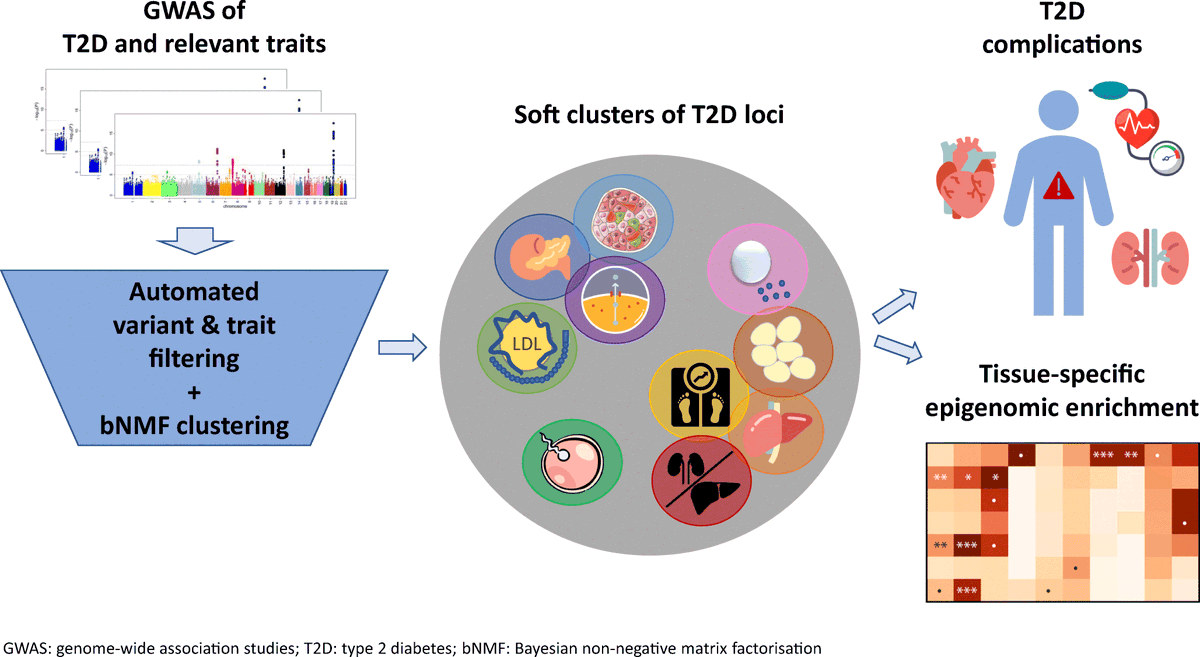
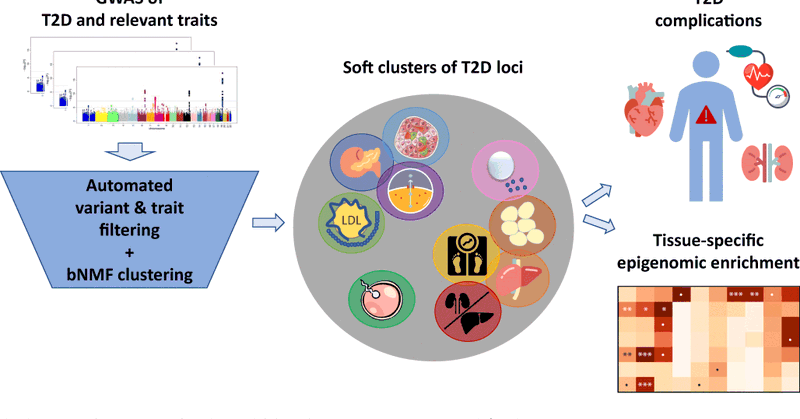
Analyses of large cohorts of patients with type 2 diabetes identified 12 disease genetic clusters with distinct cardiometabolic trait associations, providing insights on disease susceptibility across ancestries @miriam_udler @broadinstitute @CGM_MGH
0
14
41
New paper by @andywdahl and I on phenotype integration improving GWAS power while preserving specificity is now out in @NatureGenet 🎉 thank you all collaborators and reviewers!
7
63
247
My lab at @UChicago has an open postdoc position. AT #ASHG2023 AND interested in Statistical genetics and functional genomics? Please reach out and let's chat!
Postdoc fellow- Computational Biology, University of Chicago @XuanyaoLiu Come join us at #UChicago as a #bioinformatics postdoc, working on #human genetics and #functional genomics. https://t.co/VLyVm1iv2K
#ScienceJobs #job #genomics #postdoc #UC... https://t.co/VLyVm1iv2K
0
16
26
Ever wonder why people get type 2 diabetes at different BMI thresholds? So proud of this incredible effort to generate multi-ancestry T2D genetic clusters & inform on disease heterogeneity!
1/6) Type 2 diabetes (T2D) is a heterogeneous disease for which causal pathways are incompletely understood. In our new preprint, we identify 12 genetic T2D subtypes and demonstrate how they may explain variation in polygenic risk across populations
5
16
55
Second time we hold a workshop at the Schneefernerhaus, this time together with @fpcasale’s group, with a focus on statistical and machine learning methods in genomics, and invited speakers @nmancuso_ @andywdahl @TarjS Joel Mefford 😎☀️
0
2
32
Schneefernerhaus workshop 2023 done!! Thanks everyone for the wonderful talks and discussions! and we’re doing a round of Eibsee before going back to Munich, what workshop doesn’t end with a nice walk around a lake? ;) 😎☀️@andywdahl @nmancuso_ @fpcasale @TarjS
1
4
30
Our T2D genetic clustering paper is free to read in #editorspicks of @DiabetologiaJnl March issue 🧬
Up front & free to read in our March issue #editorspicks : High-throughput genetic clustering of type 2 diabetes loci reveals heterogeneous mechanistic pathways of metabolic disease https://t.co/PVo3Fexr1N
0
9
18
Our paper is featured 'Up Front' in @DiabetologiaJnl March issue!
link.springer.com
Diabetologia - Type 2 diabetes is highly polygenic and influenced by multiple biological pathways. Rapid expansion in the number of type 2 diabetes loci can be leveraged to identify such pathways....
Our editor @hinmul highlights 5 papers from our March issue - all free to read and with summaries written by the authors here https://t.co/KCvhBI4OOt 🤩
0
4
15
Ten genetic clusters of T2D identified in expanded soft clustering analysis using a high-throughput pipeline for variant and trait pre-processing @miriam_udler @hyunkyungkim_ @CGM_MGH @broadinstitute #T2D #softclustering #bNMF
https://t.co/PVo3Fexr1N
0
14
34
Interactive results are viewable on the Common Metabolic Disease Knowledge Portal @AMP_CMDKP ( https://t.co/gCYK3JjDme)
0
1
0
Huge thanks to my amazing mentor @miriam_udler and co-authors @kewesterman @JBCole150 @TimMajarian @SooHeonKwak @josep_mercader @AlisaManningPhD and others! @CGM_MGH @broadinstitute
#type2diabetes #T2D #subtypes #softclustering #bNMF #PRS
1
0
6
Thrilled to share that our manuscript on T2D genetic clustering is now published! Soft clustering analysis of type 2 diabetes loci identifies ten physiologically interpretable genetic clusters associated with distinct tissues and clinical outcomes. https://t.co/gUlJT240qb
link.springer.com
Diabetologia - Type 2 diabetes is highly polygenic and influenced by multiple biological pathways. Rapid expansion in the number of type 2 diabetes loci can be leveraged to identify such pathways....
2
17
44
Our new model of polygenic epistasis. We hope it's a helpful step toward precision medicine+understanding GWAS hits It's crazy that @dtang2000 did this as an undergrad. And very appreciative of key computational assist from @studyjeromics
super excited to share our new paper on polygenic epistasis! we introduce a new model and show how structured statistical epistasis can help us study complex trait biology. huge thanks to my friend and lab-mate @studyjeromics, and my mentor @andywdahl! https://t.co/hQ4xGPHUei
2
15
47
Are you interested in using deep learning for genomic predictions? Join my lab! please RT
5
86
186
Look for these results soon on the Common Metabolic Diseases Knowledge Portal!
High-throughput Genetic Clustering of Type 2 Diabetes Loci Reveals Heterogeneous Mechanistic Pathways of Metabolic Disease https://t.co/duplKU89Sw
#medRxiv
0
3
4
High-throughput Genetic Clustering of Type 2 Diabetes Loci Reveals Heterogeneous Mechanistic Pathways of Metabolic Disease https://t.co/duplKU89Sw
#medRxiv
0
9
32