Hyobin Kim
@HyobinKim4
Followers
92
Following
1K
Media
4
Statuses
291
Project Scientist at Cedars-Sinai Medical Center #SpatialTranscriptomics #scRNAseq #CellCellInteractions #GeneRegulation #BooleanNetworks
Los Angeles, CA, USA
Joined August 2018
Our review paper on cell-cell contact has been published in @TrendsGenetics! It was a collaborative effort with my amazing supervisor, @KJWonKJ, and my excellent colleague, .@pcnmartin, and many other wonderful contributors.
@HyobinKim4 and @pcnmartin discussed various ways to study the influence of cell-cell contact in @TrendsGenetics. It covers the proximity labeling, and algorithmic development to study cell contact from #PICseq and #SpatialTranscriptomics data.
0
2
11
RT @naturemethods: Interact-omics is a high-throughput cytometry-based framework for cellular interaction mapping. @haas_lab . https://t.co….
0
26
0
RT @KaminskiMed: 1/n .Finally out as Preprint: UNAGI, a deep generative neural network tailored to analyze time-series single-cell data, c….
0
38
0
RT @denny_zhou: Slides for my lecture “LLM Reasoning” at Stanford CS 25: Key points: .1. Reasoning in LLMs simply….
0
459
0
RT @saezlab: 🎉 The revised version of CORNETO, our unified Python framework for knowledge-driven network inference from omics data, is publ….
0
43
0
RT @LabWelch: Our paper is now in @NatureBiotech! Topological velocity inference from spatial transcriptomic data. TopoVelo infers the dire….
0
35
0
RT @Cancer_Cell: The pan-cancer proteome atlas, a mass spectrometry-based landscape for discovering tumor biology, biomarkers, and therapeu….
0
142
0
RT @mo_lotfollahi: (1/n) Can we disentangle the effects of multiple covariates (e.g., sex, age, disease) to predict multiple counterfactual….
0
18
0
RT @BoWang87: What a mind-blowing week for AI in biology!. 🚀 Xaira just dropped the largest genome-wide perturbation dataset ever last week….
0
95
0
RT @mariabrbic: Can we build multimodal models by simply aligning pretrained unimodal models with limited paired data? . We introduce STRUC….
0
36
0
RT @akshay_pachaar: Top 4 open-source LLM finetuning libraries!. From single-GPU “click-to-tune” notebooks to trillion-param clusters, thes….
0
265
0
RT @naturemethods: Out today from the Schwartz lab! A new way to study cell-cell communication from spatial transcriptomics data. Cell Neur….
0
41
0
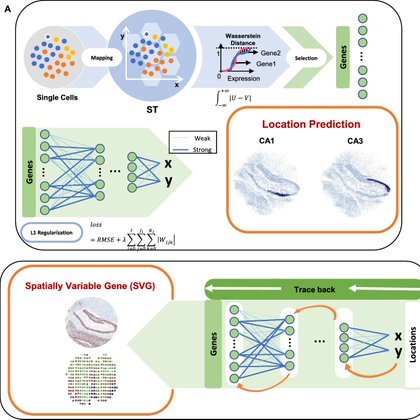
RT @KJWonKJ: SC2Spa uses a fully connected neural network (FCNN) with L1 regularization to map gene expression profiles to 2D or 3D spatia….
bmcbioinformatics.biomedcentral.com
Background Understanding cellular heterogeneity within tissues hinges on knowledge of their spatial context. However, it is still challenging to accurately map cells to their spatial coordinates....
0
1
0
RT @BoWang87: How can we make genomic foundation models actually useful to biology?! Teach them to REASON!! . 🧬 Excited to share BioReason….
0
110
0
RT @KexinHuang5: 📢 Introducing Biomni - the first general-purpose biomedical AI agent. Biomni is built on the first unified environment fo….
0
118
0
RT @naturemethods: PUPS - prediction of unseen proteins' subcellular localization - uses machine learning models to predict the localizatio….
0
18
0
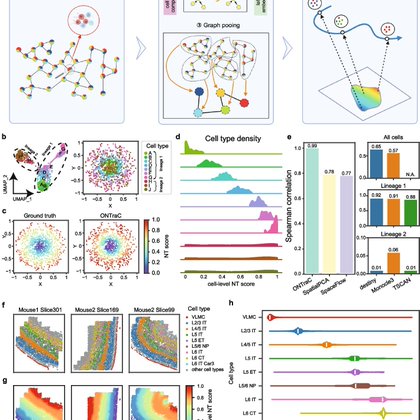
RT @gc_yuan: We are excited to announce our latest work on spatial omics data analysis has just been published! In this paper, we developed….
link.springer.com
Recent technological advances enable mapping of tissue spatial organization at single-cell resolution, but methods for analyzing spatially continuous microenvironments are still lacking. We introduce...
0
13
0
RT @LukasValihrach: From Transcripts to Cells: Dissecting Sensitivity, Signal Contamination, and Specificity in Xenium Spatial Transcriptom….
0
29
0
RT @fabian_theis: 1/ Excited to share CellFlow, a new approach for complex perturbation modeling in single-cell genomics based on flow matc….
0
71
0