FPbase
@FPbase
Followers
2K
Following
121
Media
10
Statuses
129
The Fluorescent Protein Database. tweets by @TalleyJLambert
Joined January 2019
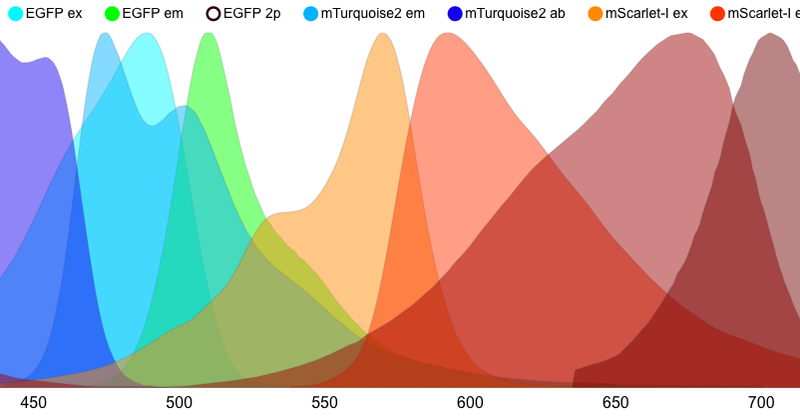
Try the new FPbase spectra viewer! Completely rewritten to be faster, more customizable, and full-featured: • SVG/PNG/CSV export • state recovery & URL sharing • keyboard shortcuts for rapid entry 💚 • efficiency calculations • full documentation https://t.co/sG8FQHD0ye
fpbase.org
An interactive fluorescence spectra viewer to evaluate the spectral properties of fluorescent proteins, organic dyes, filters, and detectors.
1
60
195
Hi Fluorescence friends! Please follow future updates on Bluesky: https://t.co/D3nuYEyGCq
0
0
1
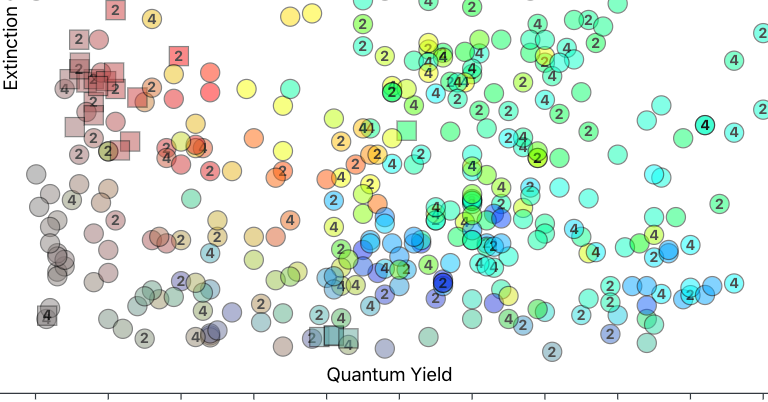
This exercise was less informative than I'd hoped, but perhaps it's interesting to someone else: spectroscopically-inferred estimates of densities of states for a selection of fluorescent proteins spanning the visible spectrum. Data from @FPbase.
1
2
5
Mutagenesis of #FbFP #LOV domain: 🟠 22 variants with 486-512 nm emission maxima 🟠 three-channel imaging via spectral unmixing 🟠 two-channel fluorescence lifetime unmixing https://t.co/5Kgc045vlh Congratulations, @ivangushchin, @Aynya5, and all the authors! @FPbase
0
1
15
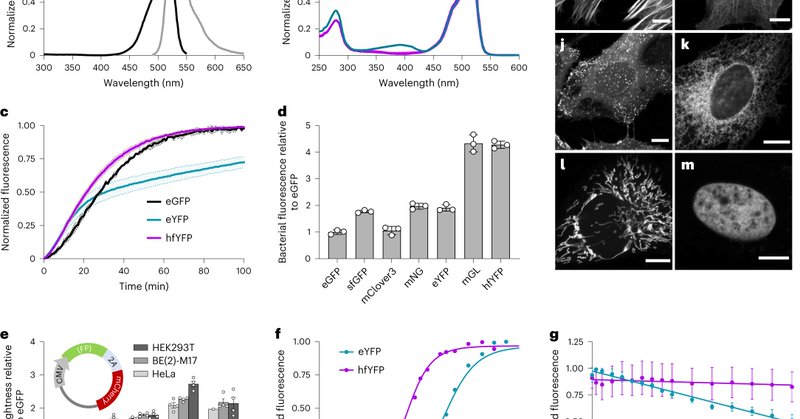
Our article reporting hyperfolder YFP (#hfYFP) and other fixative-resistant fluorescent proteins based on #mGreenLantern is now available at @naturemethods. hfYFP enhances electron- and expansion-microscopies, protein purification, and so much more! 🧵 https://t.co/hyUvgeno85
nature.com
Nature Methods - The engineered hyperfolder YFP (hfYFP) and variants offer unprecedented chemical and thermal stability, making them versatile probes for microscopy as well as for challenging...
13
124
511
Chapter on using FPbase tools out now in the MMB issue on FPs. Thank you for editing @mayankdu!
Happy to share the Methods in Molecular Biology issue on Fluorescent Proteins 🌈🎉😀. Including many “much awaited” protocols on and about Fluorescent Proteins/applications. A big THANKS to authors across the globe for contributing. 👏🏻💐 https://t.co/0yCqaDfzKw
1
5
20
this is genius: @helsinkiuni BioImaging Unit utilizes @FPbase to allow users test their fluorophores https://t.co/GGDSgRJNHq cc @Daniel_Wa19
5
3
40
anyone out there routinely express FPs in Chlamydomonas? any modern suggestions for SNR? (presumably red, but any expression gotchas?)
1
2
3
🥁 Delighted to announce that our FPCountR paper is now on bioRxiv! In this paper we describe the development of a method to allow absolute fluorescent protein quantification using microplate readers for synthetic circuit characterisation. https://t.co/RajzAm22hH 🧵
2
27
81
Always nice to see features put to use! @eszter_cs and @gbstan use the FPbase API (among other things) in a method for the calibration of fluorescence readings on microplate readers; for quantification of FP concentration https://t.co/oyotmOOiMb
biorxiv.org
This paper presents a generalisable method for the calibration of fluorescence readings on microplate readers, in order to convert arbitrary fluorescence units into absolute units. FPCountR relies on...
1
2
9
Just discovered the interactive chart of https://t.co/UwGR8SkHI6. Wow, what an amazing tool to compare different properties across a wide range of fluorescent proteins. Great work @TalleyJLambert 🙂 https://t.co/aVZ7oV6S37
fpbase.org
Plot a variety of fluorescent protein properties in different ways. Interactively change the axes of the graph and filter proteins based on certain characteristics.
1
11
52
@miCHRIScopy @TalleyJLambert FPbase is amazing! I created scope configurations for @UAZCancer core instruments, and link to it with a little QR code posted next to the scope! And made a nice lab exercise for my microscopy class. iScopeCalc rocks too. Keep ‘em coming, @TalleyJLambert!
1
1
7
Writing a methods chapter on using FPbase for an upcoming MMB book... Will include things like -Browsing/searching FPs on fpbase -Spectra viewer -microscope & reports feature If there's an aspect or detail of FPbase that you wish were better documented, let me know!
2
10
25
This is beautiful and makes me want to rewrite FPbase using it 😍
Big project, first public release! 📢 ✨ SQLModel ✨ ...the biggest thing I've built since FastAPI & Typer. 🤓 SQL DBs based on Python type hints. 😎 Each model is both a Pydantic and @sqlalchemy model. 🤯 Optimized for FastAPI 🚀 https://t.co/oEFTy828HE
1
1
3
Dear imaging companies, Please tell your customers (or make available on request) the *actual* spectra for all components (LEDs, filters, etc) in their light path. just saying "460-490nm" is not sufficient. It's amazing how many scientists can't get this info from their reps 😞
1
18
70
a reminder on the difficulty in comparing fluorescence photobleaching measurements across studies (and even across probes on the same scope) https://t.co/3yEVooVrDE
fpbase.org
FPbase is a free and open-source, community-editable database for fluorescent proteins and their properties.
0
17
27
📣 Apply for an award from Addgene to attend a conference! We're accepting applications until April 30 ☑️ Undergrad, grad student, postdoc, research tech ☑️ Working w/ fluorescent proteins ☑️ Planning to attend a conference, virtual, hybrid, or in-person https://t.co/ZmjnrYtwCy
blog.addgene.org
See the winners for the 2021 Michael Davidson and Roger Tsien Commemorative Conference Award.
1
17
26
Databases are hard... and no amount of automated error checking can catch every mistake that humans make (in the literature or in submissions). FPbase gratefully accepts help/corrections from anyone! Here's an example of what not to do if you find something needing correction 😂
2
0
23
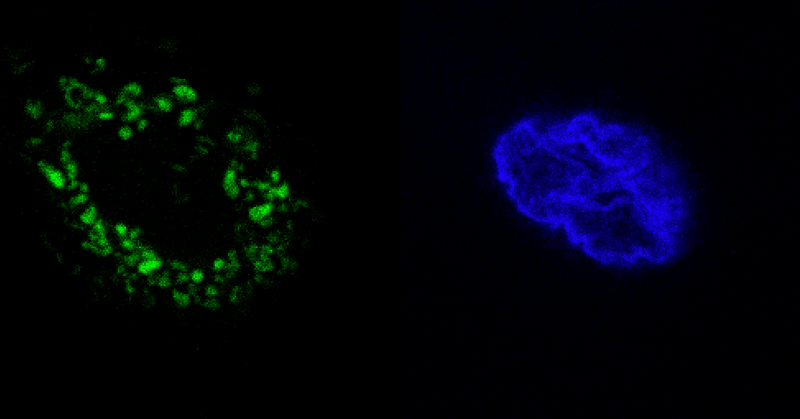
@rhodamine110 @Merz @edward_marcotte @J_A_C_S more JF spectra than you can shake a stick at. (2P coming soon) thanks @rhodamine110! https://t.co/2dhd5KVhgK
1
12
31