Ken Xie
@CuriousKX
Followers
128
Following
1K
Media
0
Statuses
445
Postdoc Fellow @IdoAmitLab. Interested in deciphering the complex cell-cell interactions in tumors
Rehovot, Israel
Joined October 2022
Excited to share our fun journey through time with the amazing team @D_Birschenkaum @FlorianIngelfi1 Yonathan @kathleenabadie @AssafWeiner from the @IdoAmitLab, measuring the temporal dynamics of immune cells in the GBM!
We are very excited to present the development of Zman-seq (“Zman”, Hebrew for “time”), the 1st technology that measures single-cell transcriptomes and physical time in vivo, led by @D_Birschenkaum, @CuriousKX, @FlorianIngelfi1, @AssafWeiner
https://t.co/pDk6ackAtV. (1/19)
4
1
22
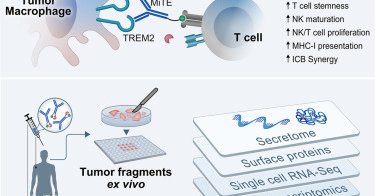
Now online! Macrophage-targeted immunocytokine leverages myeloid, T, and NK cell synergy for cancer immunotherapy
cell.com
MiTEs are myeloid-targeted immunocytokine prodrugs that block TREM2+ tumor-associated macrophages while activating cytotoxic lymphocytes via TME-specific IL-2 activity, eliciting strong anti-tumor...
0
26
76
In a new study, published in @CellCellPress, scientists from @IdoAmitLab have developed a new, dual-action strategy for cancer immunotherapy >> https://t.co/oU8ImeKXIi
1
13
33
Excited to share our study @CellCellPress introducing a novel class of macrophage-targeted immunocytokines (MiTEs) that engage myeloid, T and NK cells to boost anti-tumor immunity in solid tumors. Led by @MVLocquenghien, @PascaleZwicky & @CuriousKX (1/16) https://t.co/2z69HAdAEs
cell.com
MiTEs are myeloid-targeted immunocytokine prodrugs that block TREM2+ tumor-associated macrophages while activating cytotoxic lymphocytes via TME-specific IL-2 activity, eliciting strong anti-tumor...
25
62
202
Excited to share our latest study on MiTEs!! Really grateful to be part of this wonderful journey led by @MVLocquenghien @PascaleZwicky @IdoAmitLab
Excited to share our study @CellCellPress introducing a novel class of macrophage-targeted immunocytokines (MiTEs) that engage myeloid, T and NK cells to boost anti-tumor immunity in solid tumors. Led by @MVLocquenghien, @PascaleZwicky & @CuriousKX (1/16) https://t.co/2z69HAdAEs
0
1
9
High-plex spatial RNA imaging in one round with conventional microscopes using color-intensity barcodes - @PKU1898
https://t.co/lqj9yeq40H
0
84
392
Building on these insights, we used DIAL to titrate expression and program cell fate. Now published online at Nature Biotechnology. Links below👇 @science_sneha @mitch
🚨New paper alert!🚨 Cells integrate signals into decision-making. But how do levels of signaling influence cell-fate transitions? Is more always better? Or is there an “optimal” level for cell-fate programming? If we could tune signaling, could optimize production of neurons?
4
8
58
The Lipid Brain Atlas is out now! If you think lipids are boring and membranes are all the same, prepare to be surprised. Led by Luca Fusar Bassini with Gioele La Manno's lab, we mapped membrane lipids in the mouse brain at high resolution. https://t.co/q3AeqGsIS1
31
270
1K
1/ 🧬 MrVI just published in @NatureMethods! We’d like to take this chance to highlight a few works that have already leveraged MrVI to make scientific discoveries 🧵 📄
nature.com
Nature Methods - MrVI, based on deep generative modelling, is a unified framework for integrative, exploratory and comparative analyses of large-scale (multi-sample) single-cell RNA-seq datasets.
5
16
82
Introducing CellTransformer, a new AI tool developed with @UCSF that makes it easier to explore massive neuroscience datasets and identify important subregions of the brain. 🧵
9
146
671
Imagine a brain decoding algorithm that could generalize across different subjects and tasks. Today, we’re one step closer to achieving that vision. Introducing the flagship paper of our brain decoding program: https://t.co/lwpD2SM55G
#neuroAI #compneuro @UofT @UHN
12
99
551
Nature research paper: Proteotoxic stress response drives T cell exhaustion and immune evasion https://t.co/0afdViCh4u
nature.com
Nature - A proteotoxic stress response specific to exhausted T cells, governed by AKT signaling and accompanied by increased protein translation, represents a mechanistic vulnerability and a new...
1
44
199
In a line of foundation models for single-cell: 🚀 From CellPLM (ICLR’24, cells as tokens) → Tabula (NeurIPS’25, federated tabular FM) → now scLinguist, our first multi-omics foundation model for single-cell. Inspired by machine translation + limited paired single-cell data:
1
11
51
Core metabolic pathways Source & PDF Download Bibel, B., & The bumbling biochemist. (2024). Core Metabolic Roadmap. Zenodo. Free Download https://t.co/oHN8AuZjVU
7
176
660
Human brain #Organoid scRNAseq + scATACseq + single-cell transcription factor perturbation (CROP-seq, lentiviral gRNA library for 20 TFs, 3 gRNAs/TF, 1 gRNA detected/cell in >22k cells) in GLI3-HES1 antagonism for dorsoventral patterning in human telencephalon Pando:
0
32
208
Our Systema framework for evaluating genetic perturbation response prediction methods is now out in @NatureBiotech ✨ Predicting cellular responses to genetic perturbations is hard and current metrics often overestimate model performance. Systema helps to evaluate perturbation
Systema: a framework for evaluating genetic perturbation response prediction beyond systematic variation https://t.co/hF56Sxbj82
5
32
153
🧬🔬🧪 🎉 Our team at @czbiohub is thrilled to share TWO companion papers out today in @NatureMethods! 📦 Ultrack — robust, scalable nD cell tracking 🌐 inTRACKtive — a beautiful, open-source web viewer for lineage exploration Let’s dive in! 👇 (LINKS BELOW)
7
47
190
#InterferonPower! Thrilled for our latest work @CellCellPress! With @dfrboehmer, we dug into tons of papers & created what we hope will be a go-to resource for immunologists & non-immunologist about type I, II, III (& IV😉) #interferons! Free link 👉 https://t.co/gYwGir0npB
13
119
404
Identifying malignant cells in single-cell transcriptomics data – from first principles to computational solutions, framed around the hallmarks of cancer. New review from the lab! https://t.co/Y7mRnfDuyo
0
26
87
In the latest issue! Cross-organ metabolite production and consumption in healthy and atherogenic conditions
1
23
107
Just published in Nature (June 2025), Andersen et al. (@EAChiocca, Reardon, @QuintanaLabHMS) developed a novel viral barcode interaction-tracing (RABID-seq) platform, uncovering previously unknown immunosuppressive interactions between astrocytes and glioblastoma (GBM) cells.
2
33
176