Carmen Bravo
@Bravo_Potato
Followers
212
Following
113
Media
9
Statuses
49
Joined December 2016
💫NEW: Bravo González-Blas (@Bravo_Potato), Aerts (@steinaerts) & co uncover enhancer-gene regulatory networks underlying #hepatocyte identity and their zonation state by combining single-cell and spatial #multiomics with #DeepLearning. https://t.co/K8Qe0ovqvo
nature.com
Nature Cell Biology - Bravo González-Blas et al. uncover enhancer-gene regulatory networks underlying hepatocyte identity and their zonation state by combining single-cell and spatial...
2
4
32
🧬🔬Decoding liver mysteries with single-cell technology! The latest study by the @steinaerts lab (@_VIB_AI), published in @NatureCellBio, explores the spatial variations in hepatocyte functions. ➡️ https://t.co/JWd2VeBsLN
0
7
28
Happy to announce the first paper of my postdoc in the @TreutleinLab! A fun collaboration with @HsiuChuan_Lin where we investigated how morphogen signaling pathways can be used to increase the diversity of TF-induced neurons. Check the paper here: https://t.co/ESPXwSOIF0!
biorxiv.org
Human neurons programmed through transcription factor (TF) overexpression model neuronal differentiation and neurological diseases. However, programming specific neuron types remains challenging....
Programming diverse neuron subtypes from stem cells! Coupling patterning with proneural TF induction, we screened for 480 morphogen signaling modulations in ~700K cells. Great work with @JasperJanssens7 @TreutleinLab @GrayCampLab
https://t.co/8PGJrJdz3E
7
16
123
Exciting news! Our latest research on "Cell type directed design of synthetic enhancers" has been published in @Nature. We've harnessed deep learning to design enhancers with cell-type specificity. Find our paper here: https://t.co/wC44fDMp70. Highlights below: [1/n]
22
255
956
We are very excited to present a major breakthrough achievement – the de novo design of synthetic enhancers for selected tissues in fruit fly embryos in vivo using deep- and transfer learning, @deAlmeida_BPet al published today in @Nature
https://t.co/TIlPrBKYaK. Thread 👇(1/N)
21
198
670
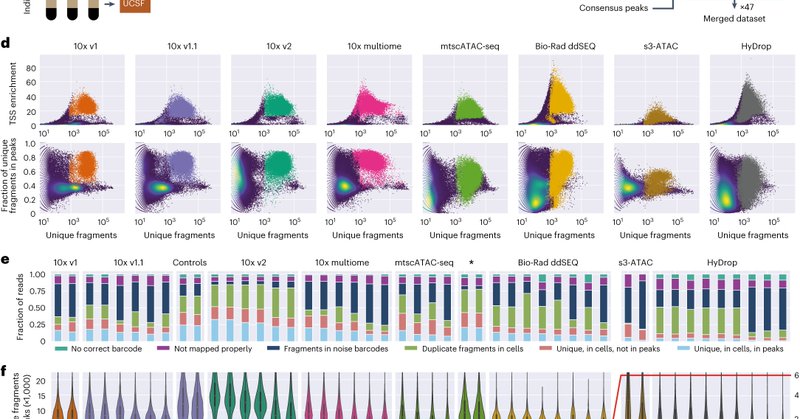
Proud to announce our systematic scATAC-seq benchmark in @NatureBiotech! The @steinaerts lab teamed up with @hoheyn and 9 other labs/companies to investigate performance differences and possible fragment biases between current scATAC-seq methods. https://t.co/A173fzdJ0a 1/n
nature.com
Nature Biotechnology - Benchmarking of scATAC-seq protocols demonstrates the effects of data quality on downstream analyses.
6
72
257
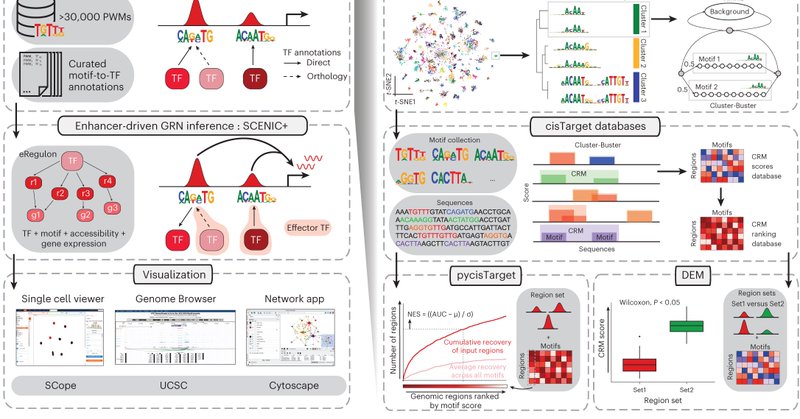
I am happy to communicate that SCENIC+ was published in @naturemethods. SCENIC+ is a workflow to infer enhancer-driven gene regulatory networks from scRNA-seq and scATAC-seq data. This work was done together with @Bravo_Potato at @steinaerts lab! 🧵1/9🧵 https://t.co/LjRdOkcuy8
nature.com
Nature Methods - SCENIC+ is a comprehensive toolbox for inferring and analyzing enhancer-driven gene regulatory networks using single-cell multiomic data.
10
147
549
Out today: SCENIC+ is a comprehensive toolbox for inferring and analyzing enhancer-driven gene regulatory networks (eGRN) using single-cell multiomic data. @steinaerts @seppe_winter @Bravo_Potato
https://t.co/favZOlwb79
nature.com
Nature Methods - SCENIC+ is a comprehensive toolbox for inferring and analyzing enhancer-driven gene regulatory networks using single-cell multiomic data.
1
193
482
And last but not least, big kudos to the amazing team that made this possible: #Hanne, @irina_M, @ibrahimihsan, #Roel, #Valerie, #Leti, @EliVerboven, #Gert, @S_Poovathingal, @zeunas, #Nikoleta, @David_Mauduit ❤️ 🧵 12/12 🧵
0
0
4
We envision that our workflow can be used as a roadmap to study other biological systems, which will further improve our understanding how cell types and their functional states are encoded in the genome. 🧵 11/12 🧵
1
0
0
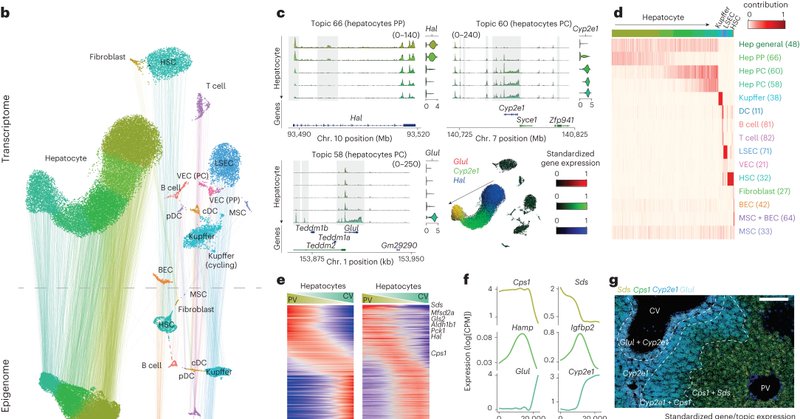
Our liver resource is available can be explored on SCope ( https://t.co/FMnxt0O29E) and the UCSC Genome Browser ( https://t.co/Geks477g2z). 🧵 10/12 🧵
1
0
1
Our data suggests that Wnt signaling orchestrates the GRN of hepatocyte zonation through repression of zonated gene expression programs by Tbx3 and Tcf7l1. 🧵 9/12 🧵
1
0
0
To assess whether this mechanism is conserved, we mapped the Tbx3 and Tcf7l1 regulons into human liver data and validated that Tbx3 motif destruction recovers enhancer activity in HepG2 🧵 8/12 🧵.
1
0
0
Taking advantage of in silico mutagenesis predictions from DeepLiver and enhancer assays, we confirmed that the destruction of Tcf7l1/2 or Tbx3 motifs in zonated enhancers abrogates their zonation bias 🧵 7/12 🧵.
1
0
0
To test these predictions further, we performed in silico Tbx3 and Tcf7l1 perturbation experiments. Our results suggest that perturbation of these TFs can switch hepatocytes zonation state 🧵 6/12 🧵.
1
0
0
We used these data to train a hierarchical deep learning model, called DeepLiver, that predicts enhancer specificity, zonation pattern and activity in hepatocytes. DeepLiver confirms that Tcf7l1/2 and Tbx3 determine zonation. 🧵 5/12 🧵
1
0
0
To uncouple enhancer accessibility and activity, we performed an in vivo MPRA on 12,000 hepatocyte enhancers, finding that 40.5% of the accessible enhancers are active. 🧵 4/12 🧵
1
0
2
Enhancer-GRN (eGRN) mapping with SCENIC+ suggests that zonation states in hepatocytes are driven by the repressors Tcf7l1 and Tbx3, which modulate the core hepatocyte GRN that is controlled by Hnf4a, Cebpa, Hnf1a, Onecut1 and Foxa1, among others. 🧵 3/12 🧵
1
0
0
We performed single-cell multiomics to identify cell types and cell states in the mouse liver. We found that cell states change in transcription and chromatin accessibility in hepatocytes, LSECs and HSC, depending on their position in the liver lobule. 🧵 2/12 🧵
1
0
1
Happy to announce that our latest work on deciphering enhancer grammar in the mouse liver is out @steinaerts #HalderLab! Link: https://t.co/ZY8WwIdFS0 🧵 1/12 🧵
biorxiv.org
Cell type identity is encoded by gene regulatory networks (GRN), in which transcription factors (TFs) bind to enhancers to regulate target gene expression. In the mammalian liver, lineage TFs have...
5
20
83