Arda Göreci
@ArdaGoreci
Followers
1K
Following
2K
Media
8
Statuses
53
AI for enzyme design | CTO at Ligo | Prev. Cell and Systems Biology @UniofOxford
San Francisco, CA
Joined August 2017
🌍 The biggest decentralized science experiment of 2025 is starting now! The protein design competition returns: we’re inviting scientists, engineers, and hackers from around the world to help design new proteins capable of neutralizing the Nipah virus, a pathogen with up to 75%
9
90
267
Today we’re releasing real-world experimental data for over 1000 novel AI-designed proteins on our new platform @proteinbase!
11
108
410
Modal is improving the quality of life for so many AI engineers and data scientists!
We're thrilled to share Modal Notebooks: a new, powerful cloud-hosted GPU notebook. It has modern real-time collaborative editing and is backed by our AI infrastructure — swap GPUs in seconds. Modal Notebooks are generally available, and you can start using them now. 🧵
0
0
1
We are hiring! I don't tend to talk about the success of @tamarindbio publicly, but we are experiencing incredible demand. Tens of thousands of scientists are regularly using the platform, and we're onboarding GPUs and people as fast as we can. We just posted multiple roles
1
10
56
It is amazing that bench scientists are being given superpowers through computational tools like Latent-X! Huge congrats to the team!!
Introducing Latent-X — our all-atom frontier AI model for protein binder design. State-of-the-art lab performance, widely accessible via the Latent Labs Platform. Free tier: https://t.co/NamdznPWjL Blog: https://t.co/2UkYDEe8a9 Technical report: https://t.co/0m2s3y7vwN
2
1
4
Super cool paper! Turns out a transformer baseline can go a long way… very excited to have Carlos on board this summer!
🌶️spicy paper alert🌶️ We present TABASCO, a new model for small molecule generation that achieves state-of-the-art PoseBusters validity while also being ~10x faster This is all achieved despite ✖️no equivariance ✖️no self-conditioning ✖️no bond modelling what we found🧵(1/n)
2
0
3
This is absolutely amazing! The most interesting part of this breakthrough for me is how many people were saying “we don’t have enough antibody data” - well, the @chaidiscovery team have shown that we might have enough to solve the problem.
We’re excited to introduce Chai-2, a major breakthrough in molecular design. Chai-2 enables zero-shot antibody discovery in a 24-well plate, exceeding previous SOTA by >100x. Thread👇
2
1
8
Beyond thrilled to announce that @JohnJumperSci Nobel prize winner and Distinguished scientist from Google DeepMind will be joining our event as a panelist!
Excited to announce the AI x Bio “unconference” hosted by Ligo Biosciences and FutureHouse on June 18! Featuring Simon Kohl, Sam Rodriques, Brandon Wood, Nima Alidoust.
0
1
5
Excited to announce the AI x Bio “unconference” hosted by Ligo Biosciences and FutureHouse on June 18! Featuring @saakohl , @SGRodriques , @bwood_m , @nalidoust.
0
5
11
This dataset will turbocharge deep learning methods trained on structural data! Doubling the PDB protein-ligand interactions in 18 months🔥
Really pleased to share what I have been working on for 2 months: 🇬🇧 UK SovAI are today announcing our £8m seed investment into OpenBind - A consortium that will actually make AI for drug discovery great by generating 500k experiment protein-ligand complexes!! Explainer 🧵 (1/n)
0
1
3
Excited to announce the AI x Bio “unconference” hosted by Ligo Biosciences and FutureHouse on June 18! Featuring Simon Kohl, Sam Rodriques, Brandon Wood, Nima Alidoust.
2
3
13
This is one of the best use-cases of AI in biotech I’ve ever seen. Brandon and the Axiom team collected paired molecule-cell image data and trained an AI model to learn drug toxicity determinants. It’s embeddings became more predictive than a $15k / molecule 3D liver cell assay.
Announcing Axiom: Eliminating drug toxicity, without using animals! Alex Beatson and I founded Axiom a little over over a year ago with the mission of eliminating drug toxicity by replacing traditional experiments, such as animal testing, with AI models. We are excited to
0
0
3
Today, we're launching Orpheus Multilingual, a family of open-source models that makes state-of-the-art TTS accessible to billions of new people! 🌎🌎 (1/5)
46
99
876
This is awesome!! AF3 fully open-source. Huge congratulations to the team. This is going to completely change structural biology / protein design workflows.
Thrilled to announce Boltz-1, the first open-source and commercially available model to achieve AlphaFold3-level accuracy on biomolecular structure prediction! An exciting collaboration with @jeremyWohlwend, @pas_saro and an amazing team at MIT and Genesis Therapeutics. A thread!
1
3
29
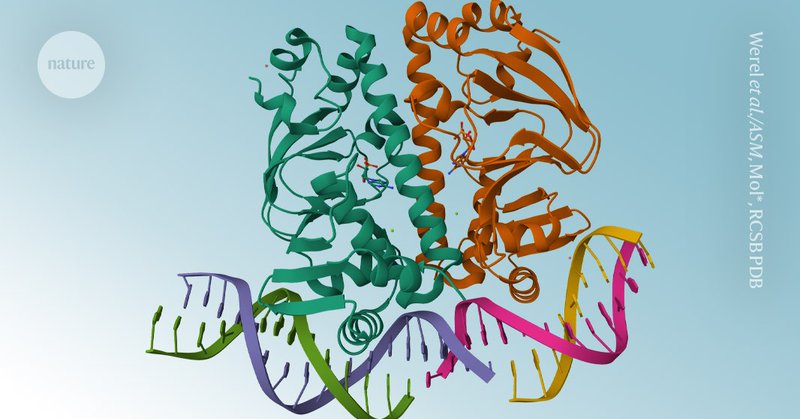
Disappointing to see @Nature fail to acknowledge that all teams who reproduced DeepMind's code (Ligo Biosciences, @chaidiscovery, @Baidu_Inc, ByteDance) found significant errors in the pseudocode which prevented training and compromised reproducibility.
nature.com
Nature - The code underlying the Nobel-prize-winning tool for modelling protein structures can now be downloaded by academics.
1
5
15
Excited for Ligo Biosciences to be included in both @Nature Magazine and @ScienceMagazine in their recent articles on the release of @GoogleDeepMind 's AlphaFold3 code.
science.org
Six months after backlash, AI company fulfills pledge to make AlphaFold3’s full computer model available for noncommercial use
1
15
74
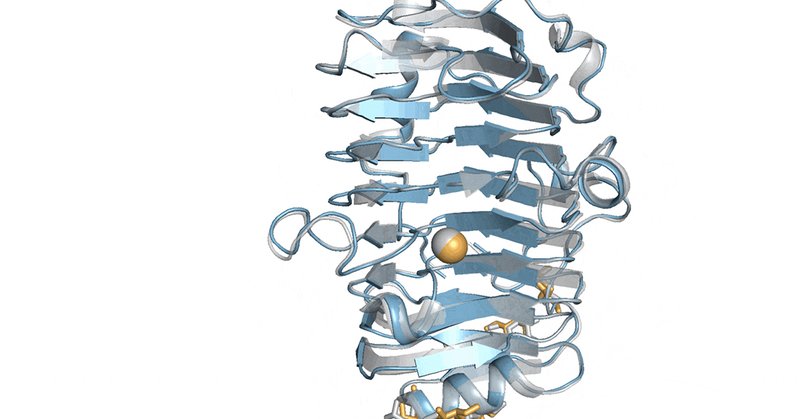
Alex wrote a Triton kernel that allows the model to scale to thousands of residues! Runs faster and is more memory efficient. Huge contribution, check it out on the repo!
Had the pleasure of building an efficient Triton kernel to help scale @ArdaGoreci Ligo's open-source AlphaFold3 model! A new component in AlphaFold3 is the MSA pair weighted averaging algo -- our Triton implementation is 3x faster with 10x less peak memory usage! (1/3)
0
4
30
DeepMind mentions 4 “homogeneous blocks,” - it is ambigous if these are shared weights or just have the same architecture. If shared, the layers will have grads from the first 3, but should skip final MSA stack. In AlphaFold2 the paper was more explicit about “shared weights”.
0
0
1
Huge thanks to Matthew Clark for his animation. Check out his awesome work here: https://t.co/4JrVvyFugC. He did a slowed down version as well!
2
5
32