Anat Kreimer 💔
@AnatKreimer
Followers
128
Following
133
Media
1
Statuses
58
Assistant Prof. at Rutgers University @RutgersCABM mainly focused on Computational Functional Genomics.
Joined October 2013
🚀 Proud to launch ⭐Path2Space⭐ — the first AI to discover spatial biomarkers for cancer treatment! It predicts spatial gene expression from histopathology, enabling large-scale tumor microenvironment profiling. https://t.co/kYFfWEYqyi 🧵1/11
11
112
334
Our paper is out today !!! https://t.co/bpjB3o4fiO
@RutgersCABM @NadavAhituv @nir_yosef1 @TakaInoue5 @TalAshuach
link.springer.com
Background Increasing evidence suggests that a substantial proportion of disease-associated mutations occur in enhancers, regions of non-coding DNA essential to gene regulation. Understanding the...
0
1
6
Massively parallel jumping assay (MPJA). High throughput assay to assess the jumping potential of thousands of retrotransposon sequences/haplotypes. Amazing work by @navneetkmatharu, @JJZhao1203, @KircherLab and many others.
biorxiv.org
The human genome contains millions of retrotransposons, several of which could become active due to somatic mutations having phenotypic consequences, including disease. However, it is not thoroughly...
1
11
49
Happy to share our paper is published in NAR today !!! @NadavAhituv @YosefLab @TalAshuach @TakaInoue5 @RutgersCABM
https://t.co/wjjelW2mcc
academic.oup.com
Abstract. The advent of perturbation-based massively parallel reporter assays (MPRAs) technique has facilitated the delineation of the roles of non-coding
0
2
12
I usually leave politics off this site. But the amount of antisemitism on display is pretty crazy. To all my Jewish friends and colleagues, I’m sorry and realizing I really didn’t understand/see it until now.
245
509
4K
Our joint work with @lab_berger (@ekimbarisc) and Ron Shamir (David Pellow, @Bio_Lian) on minimizers orders based of minimum decycling sets to speed up high-throughput sequencing applications is finally published in @genomeresearch: https://t.co/h8nT9ZavyO. Thanks @usisraelbsf!
Great job @Bio_Lian presenting our work on minimum-decycling-set-based minimizer orders! #RECOMB2023 @RECOMBconf
0
3
13
It was a pleasure to host @AnatKreimer and hear about her work on multi-parallel reporter assays (MPRAs) to decipher the function of non-coding variants and their association with diseases @ubarilan. Thanks for visiting and enjoy your time in Israel!
0
1
11
New manuscript with exciting findings 🥳 Utilizing nullomers in cell-free RNA for early cancer detection
medrxiv.org
Early detection of cancer can significantly improve patient outcomes; however, sensitive and highly specific biomarkers for cancer detection are currently missing. Nullomers are short sequences that...
1
3
15
MPRA on 680,000 sequences comprehensively covering all active regulatory elements in 3 cell lines (HepG2,K562,iPSCs). Tour de force by @vagar112 & @TakaInoue5, @JShendure and @KircherLab as part of our @ENCODE_NIH project. https://t.co/uqkvzuVFNi
biorxiv.org
The human genome contains millions of candidate cis -regulatory elements (CREs) with cell-type-specific activities that shape both health and myriad disease states. However, we lack a functional...
2
35
137
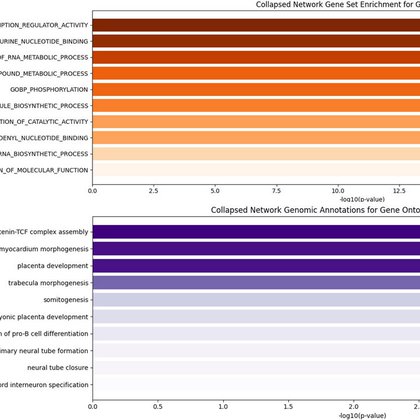
MPRA identifies 46,802 enhancers & 164 psychiatric disorder variants in the developing cortex & organoids. Outstanding work by Chengyu Deng, @seawhalen & Katie Pollard in collaboration with @LabNowakowski, @brainevodevo + many others in psychENCODE. https://t.co/znS2L2jfEg
biorxiv.org
Nucleotide changes in gene regulatory elements are important determinants of neuronal development and disease. Using massively parallel reporter assays in primary human cells from mid-gestation...
1
32
118
Excited and proud to present the first paper from the Kreimer lab!!! https://t.co/Na5oEw5a5u Great work by Justin Koesterich and collaboration with Stephan Sanders @joonomics
@TakaInoue5
@NadavAhituv
@RutgersCABM
mdpi.com
Autism spectrum disorder (ASD) is a common, complex, and highly heritable condition with contributions from both common and rare genetic variations. While disruptive, rare variants in protein-coding...
3
4
19
Extremely honored for this. Thanks to my lab, IHG members and staff and so many people at UCSF! Looking forward to advancing the IHG and genetics and genomics research at UCSF!
38
21
249
Machine learning dissection of human accelerated regions in primate neurodevelopment
cell.com
Whalen et al. couple deep learning with functional assays in chimpanzee and human cells to interrogate the neurodevelopmental enhancer potential of 2,645 human accelerated regions (HARs). Activity is...
1
4
27