Akshay Subramanian
@AkshaySubraman9
Followers
347
Following
1K
Media
5
Statuses
127
PhD student @ MIT | Generative models and atomistic simulations for molecules and materials | Personal website: https://t.co/eyZwjj9lhy
Cambridge, MA
Joined February 2019
Out now in @ChemicalScience! Give it a read if you're interested in molecular design, synthesizability, and/or organic electronics! Co-authors: James Damewood, @junonam_ , @kevinpgreenman , @Avni_Singhal , and @RGBLabMIT. https://t.co/gUwiD3TSLs
pubs.rsc.org
Organic optoelectronic materials are a promising avenue for next-generation electronic devices due to their solution processability, mechanical flexibility, and tunable electronic properties. In...
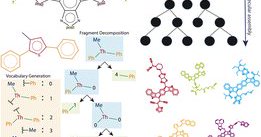
📢New preprint out! We constrain the molecular generation space to follow the "symmetry" of patented molecules that are likely to be synthesizable. Achieved with "symmetry-aware" fragment decomposition, and a constrained Monte Carlo Tree Search generator. https://t.co/NWidW2Wx9y
0
4
19
@max_simchowitz Overgeneralisation is probably the main reason why we're still so reliant on it. I think we are also intentionally collapsing the distribution towards its more canonical examples (guidance is just one way to do that, various forms of post-training are also used for this). The
2
6
104
One of my long-standing dreams is to one day create a room temperature superconductor - asked Nano Banana Pro to help visualise the kind of future this would enable! 🚀
6
7
125
Scientific foundation models are converging to a universal representation of matter. Come chat with us at #NeurIPS! We (@SoojungYang2 @RGBLabMIT) have an oral spotlight at the #NeurIPS #UniReps workshop and will also poster at #AI4Mat. 🖇️: https://t.co/H8ZRlpvJUj 🧵(1/5)
1
3
18
Today, we present a step-change in robotic AI @sundayrobotics. Introducing ACT-1: A frontier robot foundation model trained on zero robot data. - Ultra long-horizon tasks - Zero-shot generalization - Advanced dexterity 🧵->
433
662
5K
(1/n) Can diffusion models simulate molecular dynamics instead of generating independent samples? In our NeurIPS2025 paper, we train energy-based diffusion models that can do both: - Generate independent samples - Learn the underlying potential 𝑼 🧵👇 https://t.co/TSurVY3YEl
12
139
837
Tired to go back to the original papers again and again? Our monograph: a systematic and fundamental recipe you can rely on! 📘 We’re excited to release 《The Principles of Diffusion Models》— with @DrYangSong, @gimdong58085414, @mittu1204, and @StefanoErmon. It traces the core
50
465
2K
My pleasure to come on Dwarkesh last week, I thought the questions and conversation were really good. I re-watched the pod just now too. First of all, yes I know, and I'm sorry that I speak so fast :). It's to my detriment because sometimes my speaking thread out-executes my
The @karpathy interview 0:00:00 – AGI is still a decade away 0:30:33 – LLM cognitive deficits 0:40:53 – RL is terrible 0:50:26 – How do humans learn? 1:07:13 – AGI will blend into 2% GDP growth 1:18:24 – ASI 1:33:38 – Evolution of intelligence & culture 1:43:43 - Why self
587
2K
17K
We are building AI scientists and the autonomous laboratories for them to operate. Our goal is to accelerate science. Incredibly excited to be a part of this team. Read more about us @periodiclabs, on our website and in this NYT article.
Today, @ekindogus and I are excited to introduce @periodiclabs. Our goal is to create an AI scientist. Science works by conjecturing how the world might be, running experiments, and learning from the results. Intelligence is necessary, but not sufficient. New knowledge is
4
18
97
As de novo pMHC binders become commodity, the role of the wild type TCR starts to evolve. Both have their place, but lately it feels like all progress was in binders. No longer. Pleased to report that the first AI-TCR against a non-viral epitope has been achieved TCR-TRANSLATE🧵
4
13
43
People powered. Platform enabled. Progress delivered. This summer, our #MachineLearning interns helped make our biology even more programmable—advancing de novo protein design, antibody engineering, and automated high-throughput cryoEM. At our annual #ML Internship Symposium,
0
2
26
🚀 Come check our poster at ICML @genbio_workshop! We show that pretrained MLIPs can accelerate training of Boltzmann emulators — by aligning their internal representations. Coauthors @LucasPinede, @junonam_, @RGBLabMIT (1/n)
2
17
150
Happy to have contributed to this awesome project!
Today in the journal Science: BioEmu from Microsoft Research AI for Science. This generative deep learning method emulates protein equilibrium ensembles – key for understanding protein function at scale. https://t.co/WwKjj5B0eb
1
7
86
Excited to share that I'll be interning with @generate_biomed this Summer, where I'll be working on generative models for proteins! 🧬
0
0
12
Excited to start my research internship this Summer + Fall at @RealityLabs where I'll be working on vision-language models for robotics! If you're in the Seattle-area and would like to meet up, let me know!
4
4
102
I want to share my latest (very short) blog post: "Active Learning vs. Data Filtering: Selection vs. Rejection." What is the fundamental difference between active learning and data filtering? Well, obviously, the difference is that: 1/11
14
76
563
Really happy to have been on the team while this was going on. Also excited about what it means for sampling! Check out the blog post about our work sampling from these models:
linkedin.com
Announcing the newest releases from Meta FAIR. We’re releasing new groundbreaking models, benchmarks, and datasets that will transform the way researchers approach molecular property prediction,...
Excited to share our latest releases to the FAIR Chemistry’s family of open datasets and models: OMol25 and UMA! @AIatMeta @OpenCatalyst OMol25: https://t.co/UhIDiFFzzN UMA: https://t.co/eQ2wPIxbxY Blog: https://t.co/VFaeynyZN7 Demo: https://t.co/Dj29ZfhBRO
0
1
22
Out now in Journal of Vision! Measuring inference-time compute capability in neural networks (and humans) before it was cool 😎 https://t.co/FQNl2QWI1e
Excited to share my first project as a PhD student! To bring 'time' into model-human comparison, we present a large dataset of timed object recognition and test the ability of dynamic NNs to display a human-like speed-accuracy tradeoff. https://t.co/2eMR0u8h4P Summary: (1/5)
0
6
31
Thrilled to share my recent work with @sokrypton, @grocklin, @KotaroTsuboyama, and @JustasDauparas: https://t.co/ixsbgB7M8L (1/9)
biorxiv.org
Generative protein modeling provides advanced tools for designing diverse protein sequences and structures. However, accurately modeling the conformational landscape and designing sequences—ensuring...
2
23
123
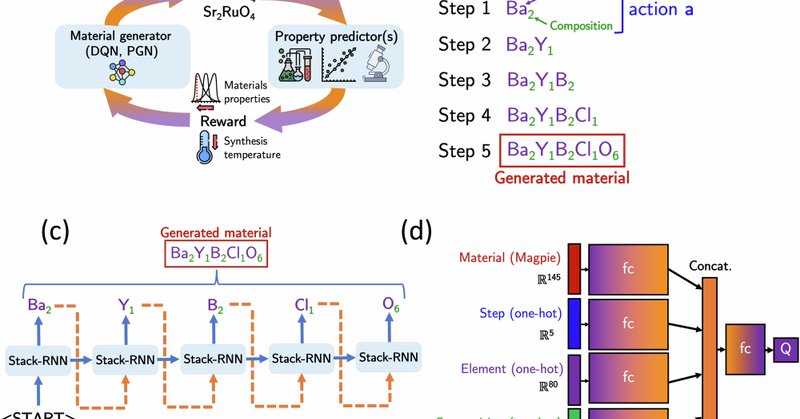
Our paper on RL for proposing inorganic materials is now published in npj Computational Materials. We compare policy- and value-based RL approaches, and demonstrate their applications in multi-objective optimization to identify promising candidates. https://t.co/V2GWVwnLAm
nature.com
npj Computational Materials - Deep reinforcement learning for inverse inorganic materials design
0
1
14