Soojung Yang
@SoojungYang2
Followers
960
Following
1K
Media
3
Statuses
202
AI+simulation for protein dynamics! PhD student @ MIT @RGBLabMIT / prev intern @MSFTResearch / website: https://t.co/xHAeCEbNpO
Joined September 2020
Check out our new preprint on converging representations in models of matter! https://t.co/gVmHlddq6e TLDR: Chemical foundation models (e.g., MLIPs) with very different architectures/tasks become increasingly **internally aligned** as they improve. See Sathya’s visual summary 👇
arxiv.org
Machine learning models of vastly different modalities and architectures are being trained to predict the behavior of molecules, materials, and proteins. However, it remains unclear whether they...
Scientific foundation models are converging to a universal representation of matter. Come chat with us at #NeurIPS! We (@SoojungYang2 @RGBLabMIT) have an oral spotlight at the #NeurIPS #UniReps workshop and will also poster at #AI4Mat. 🖇️: https://t.co/H8ZRlpvJUj 🧵(1/5)
1
1
17
@unireps @sathyaedamadaka And posters, @ SPIGM "Bayesian Optimization for Biochemical Discovery with LLMs" @ AI4Mat "Universally Converging Representations of Matter Across Scientific Foundation Models" Hit me up for coffee chat!
0
0
0
NeurIPS⛵️ Find me here on 12/6! Oral @unireps, S1: "Universally Converging Representations of Matter Across Scientific Foundation Models" @sathyaedamadaka will present our work! Oral @AI4Mat, 4:10PM: "Guided Diffusion for A Priori Transition State Sampling" I'll be presenting.
2
1
12
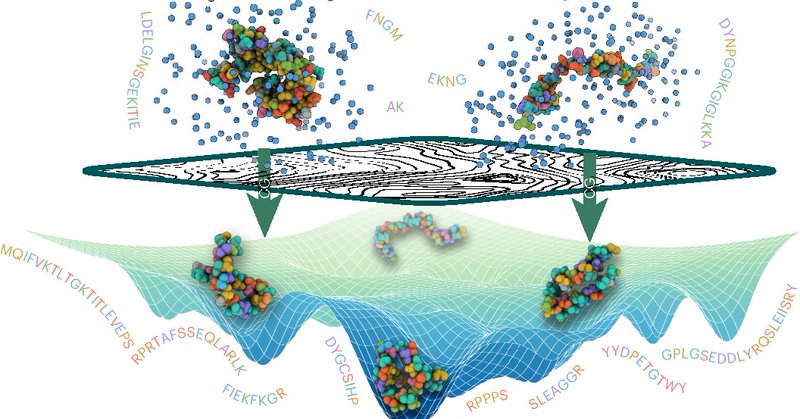
This motivated my 2024 work ( https://t.co/YreWI84Wch), where I showed that models across both input modalities strongly align and share similar statistical fingerprints (e.g., cysteines are consistently encoded with the lowest intrinsic dimensions).
openreview.net
Protein foundation models produce embeddings that are valuable for various downstream tasks, yet the structure and information content of these embeddings remain poorly understood, particularly in...
1
1
3
@sathyaedamadaka It’s time to look more closely at what foundation models are actually learning internally. Representation will be the key to transferring, integrating, and interpreting knowledge across scientific foundation models. I’m heading to NeurIPS workshops. Excited for more discussion🤩
0
1
2
@sathyaedamadaka It’s time to look more closely at what foundation models are actually learning internally. Representation will be the key to transferring, integrating, and interpreting knowledge across scientific foundation models. I’m heading to NeurIPS workshops. Excited for more discussion🤩
0
1
2
Now with @sathyaedamadaka, we took this much further in one year. We examined 59 chemical models spanning molecules, polymers, and crystalline materials, and found even stronger evidence that models with vastly different architectures/tasks converge to aligned representations.
1
0
2
This motivated my 2024 work ( https://t.co/YreWI84Wch), where I showed that models across both input modalities strongly align and share similar statistical fingerprints (e.g., cysteines are consistently encoded with the lowest intrinsic dimensions).
openreview.net
Protein foundation models produce embeddings that are valuable for various downstream tasks, yet the structure and information content of these embeddings remain poorly understood, particularly in...
1
1
3
In the protein community, we’ve known that scaling up protein language models allows structural information to emerge from evolutionary sequence data. That led me to wonder: what if protein language and structure models are converging to the same internal statistical space?
1
0
2
looking forward to being in Boston this week & hope to see you at BPDMC 🤗🔬
We can't wait until Wednesday, November 19th 2025 to arrive because we have @ginaelnesr from @PossuHuangLab speaking at 7pm EST in Room 6055, Longwood Center, @DanaFarber "Learning millisecond protein dynamics from what is missing in NMR spectra" https://t.co/E8bDyGzQgJ
0
4
22
(1/n) Can diffusion models simulate molecular dynamics instead of generating independent samples? In our NeurIPS2025 paper, we train energy-based diffusion models that can do both: - Generate independent samples - Learn the underlying potential 𝑼 🧵👇 https://t.co/TSurVY3YEl
12
141
838
Thank you UW–Madison for this great honor! In 2016 I changed my group to AI4Science and went all-in on deep learning solutions for the molecular sciences. Best decision of my career in hindsight. Amazing what's become of that field - now AI for Science is a @MSFTResearch lab.
Congratulations to Frank Noé, Partner Research Manager at Microsoft Research AI for Science, who received the 2025–2026 Joseph O. Hirschfelder Prize in Theoretical Chemistry for pioneering contributions at the interface of AI and chemistry. https://t.co/FMCPmbfAW1
9
5
65
Want to join our efforts @MSFTResearch AI for Science to push the frontier of AI for materials? We are the team behind MatterGen & MatterSim and we have 2 job openings! Each can be in Amsterdam, NL, Berlin, DE, or Cambridge, UK. It is a rare opportunity to join a highly talented,
0
24
77
Our development of machine-learned transferable coarse-grained models in now on Nat Chem! https://t.co/HGngd8Vpop I am so proud of my group for this work! Particularly first authors Nick Charron, Klara Bonneau, @sayeg84, Andrea Guljas.
nature.com
Nature Chemistry - The development of a universal protein coarse-grained model has been a long-standing challenge. A coarse-grained model with chemical transferability has now been developed by...
3
40
128
@genbio_workshop @LucasPinede @junonam_ @RGBLabMIT @sihyun_yu Unfortunately both @LucasPinede and I won't be at ICML in person this year. But our poster will be at the workshop 😂 Feel free to DM us or email our lead author (lucasp at https://t.co/Zjbm3tdReJ) about the paper! Preprint coming very soon, please tune in! (4/n, N=4)
0
0
3
@genbio_workshop @LucasPinede @junonam_ @RGBLabMIT At low noise, emulator scores ≈ physical forces. Even w/o alignment loss, emulator embeddings drift toward MLIP's. Inspired by @sihyun_yu’s REPA, we add a loss to encourage this — and training becomes faster & better. (3/n)
1
0
3
@genbio_workshop @LucasPinede @junonam_ @RGBLabMIT Boltzmann emulators can sample conformations cheaper than MD but are expensive to train. They need equilibrium data, which is very hard to get. But… MLIPs like MACE are trained on tons of noneq. data. What if we transfer knowledge from force models to generative models? (2/n)
1
0
1
🚀 Come check our poster at ICML @genbio_workshop! We show that pretrained MLIPs can accelerate training of Boltzmann emulators — by aligning their internal representations. Coauthors @LucasPinede, @junonam_, @RGBLabMIT (1/n)
2
17
151
Understanding protein motion is essential to understanding biology and advancing drug discovery. Today we’re introducing BioEmu, an AI system that emulates the structural ensembles proteins adopt, delivering insights in hours that would otherwise require years of simulation.
Today in the journal Science: BioEmu from Microsoft Research AI for Science. This generative deep learning method emulates protein equilibrium ensembles – key for understanding protein function at scale. https://t.co/WwKjj5B0eb
164
419
3K
Happy to have contributed to this awesome project!
Today in the journal Science: BioEmu from Microsoft Research AI for Science. This generative deep learning method emulates protein equilibrium ensembles – key for understanding protein function at scale. https://t.co/WwKjj5B0eb
1
7
86