Yang Zhang
@zocean636
Followers
184
Following
503
Media
0
Statuses
68
Joined August 2015
RT @jmuiuc: Final version of our paper @eLife (led by @belmont_andrew): Major nuclear locales define nuclear genome organization and functi….
elifesciences.org
New insights into nuclear genome organization were generated by comparing genome localization relative to three major nuclear locales- nuclear lamina, nuclear speckles, and nucleoli- across several...
0
8
0
RT @naturemethods: A new benchmarking study assesses methods for comparing chromatin contact maps in 3D genome research. @GjoniKatie, @Evon….
0
12
0
RT @jmuiuc: Proper and meaningful benchmark datasets are crucial for advancing genomic LLMs/FMs, and ML methods for genomics in general. Fa….
0
6
0
RT @belmont_andrew: Genome-wide analyses correlate nuclear speckle proximity with gene expression. We share our minireview discussing poss….
0
9
0
RT @james_y_zou: Very excited to share our primer on LLM for biology @naturemethods! We discuss how to use:.⚡️human & protein LMs (eg #ESM)….
0
65
0
RT @michaelkosicki: Beta-version of the new VISTA Enhancer Browser, 7 years in the making, with support for developmental stages, different….
0
53
0
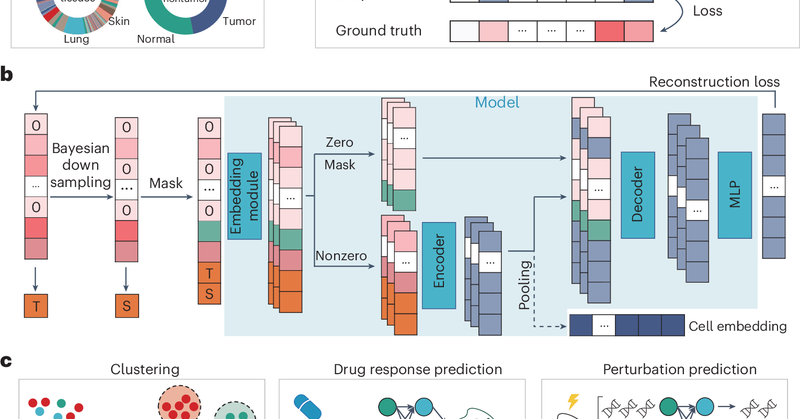
RT @XuegongZ: Our scFoundation has been finally published. It is currently one of the largest cellular models, pretrained on the transcript….
nature.com
Nature Methods - scFoundation, with 100 million parameters covering about 20,000 genes, pretrained on over 50 million single-cell transcriptomics profiles, is a foundation model for diverse tasks...
0
39
0
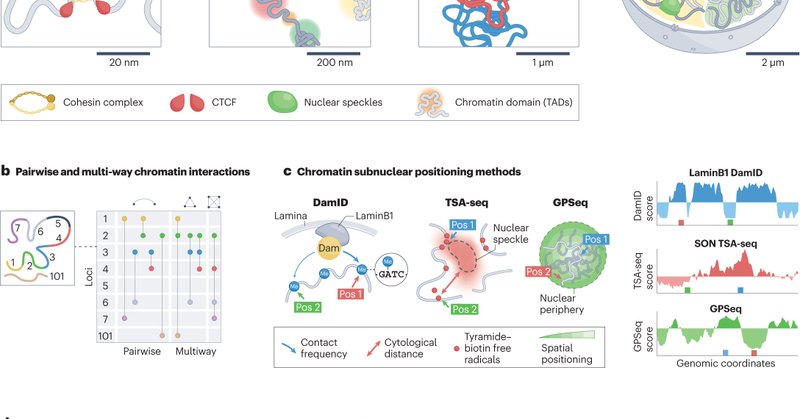
RT @belmont_andrew: Excited to share new results from our 4DN collaboration between the Gilbert, Ma, van Steensel, and Belmont research gro….
biorxiv.org
Models of nuclear genome organization often propose a binary division into active versus inactive compartments yet typically overlook nuclear bodies. Here we integrated analysis of sequencing and...
0
19
0
RT @naturemethods: scGHOST offers a computational tool to annotate single cell subcompartments from scHi-C or imaging data using graph-base….
0
5
0
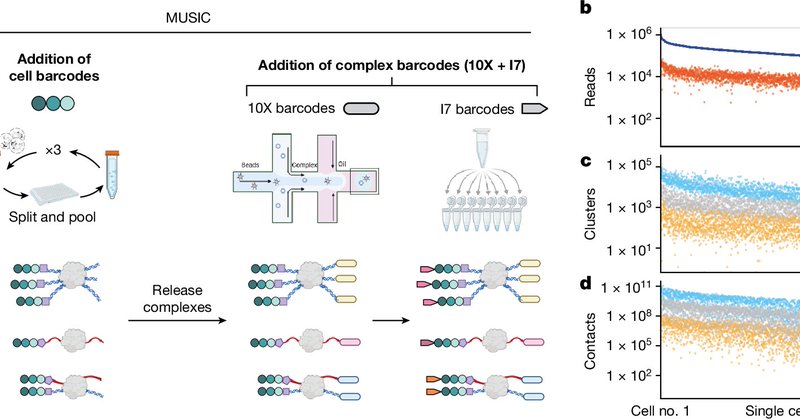
RT @Irene_xzwen: Thrilled to announce our paper in @Nature introducing MUSIC! For the first time, MUSIC enables simultaneous mapping of mul….
nature.com
Nature - We introduce multinucleic acid interaction mapping in single cells (MUSIC), for concurrent profiling of multiplex chromatin interactions, gene expression and RNA–chromatin...
0
87
0
RT @anshulkundaje: Great application of DNA language models to prokaryotic genomes. Excellent impactful application use cases of the model.….
0
56
0
RT @apombo1: Check out the upcoming Cold Spring Harbor meeting on Genome Organization & Nuclear Function, Apr 20 – May 4, 2024, on advances….
0
78
0
RT @Wings7Spread: Computational genomics Postdoc & PhD openings @ Duke🧑💻🧬🥰 Postings: | ; L….
yi-zhang-compbio-lab.github.io
Yi Zhang’s computational biology lab at Duke University (2024)
0
31
0
RT @belmont_andrew: Pleased to share our TSA-seq mapping of human genome localization relative to nucleoli and pericentric heterochromatin….
0
8
0
RT @jmuiuc: In @NatureRevGenet today, we discuss recent advances in computational methods for #3Dgenome analysis, pointing to an integrativ….
nature.com
Nature Reviews Genetics - In this Review, Zhang et al. discuss how recent advances in computational methods are helping to reveal the multiscale features involved in genome folding within the...
0
70
0
RT @NatureRevGenet: Computational methods for analysing multiscale 3D genome organization #Review by @zocean636, @L….
nature.com
Nature Reviews Genetics - In this Review, Zhang et al. discuss how recent advances in computational methods are helping to reveal the multiscale features involved in genome folding within the...
0
59
0
RT @SCSatCMU: Yang Zhang, a project scientist in the @CMUCompBio, received a $25,000 grant from the @ChanZuckerberg Initiative for his proj….
cs.cmu.edu
Yang Zhang, a project scientist in the Computational Biology Department, received a $25,000 grant from the Chan Zuckerberg Initiative (CZI) for his project "Napari-Nucleome: Integrative Multi-Omic...
0
2
0
@cziscience @jmuiuc @nimezhu Our project napari-nucleome ( aims to enable users of napari to perform interactive exploration of multi-modal datasets, including imaging, multi-omics and 3D genome structure models for studying the cell nucleus.
chanzuckerberg.com
0
0
7