Tobias Marschall
@tobiasmarschal
Followers
1K
Following
573
Media
12
Statuses
379
Algorithms, bioinformatics, genomics, and medical data science at @HHU_de / @MedHHU / @UniklinikDUS. Own views.
Düsseldorf, Germany
Joined January 2018
Thrilled to announce the largest long-read based SV resource on open samples (1kGP) to date. Highlights include a pangenome augmentation pipeline, sequence resolution of 74k insertions, fine-scale SV classification and evidence for recurrent SV formation. Great team effort:.
Long-read sequencing and structural variant characterization in 1,019 samples from the 1000 Genomes Project #biorxiv_genomic.
2
32
96
RT @NicoChatron: Looking for the best way to “complete” day3 of #eshg2024 and prepare the party 🎉 🕺 💃! Come and join @AlexReymondESHG and I….
0
2
0
RT @ewanbirney: Heading home from #bog24 on the overday flight (sorry to miss the last session) and here are some thoughts from this Biolog….
0
26
0
RT @TimofeyProdanov: We finally uploaded a preprint on Locityper—targeted genotyper for complex genes, on which @tobiasmarschal and I worke….
biorxiv.org
The human genome contains numerous structurally-variable polymorphic loci, including several hundred disease-associated genes, almost inaccessible for accurate variant calling. Here we present...
0
5
0
RT @GenomeWeb: Long-Read Nanopore Sequencing Boosts Structural Variant Analysis in 1000 Genomes Project Dataset
genomeweb.com
Two research teams have been reanalyzing 1000 Genomes Project samples with nanopore long reads to build a more comprehensive variant catalog.
0
16
0
@schloissnig @samarendra89827 @BerniRdgz @janaebler @carsten_hain @Celialou5 @asoylev, Patrick Hüther, @HufsahA @TimofeyProdanov, Mila Asparuhova, Sarah Hunt, @tobias_757, @JanKorbel5.
0
2
2
RT @glennis_logsdon: I'm absolutely thrilled to share that our work on human centromere variation and evolution has been published in @Natu….
0
136
0
RT @sergeynurk: One more opening in my group at @nanopore. This time in our New York office! .
ejnh.fa.em2.oraclecloud.com
We are looking for a motivated individual with programming expertise to help us design and implement new bioinformatics tools, libraries, and modules for various applications of Oxford Nanopore...
0
38
0
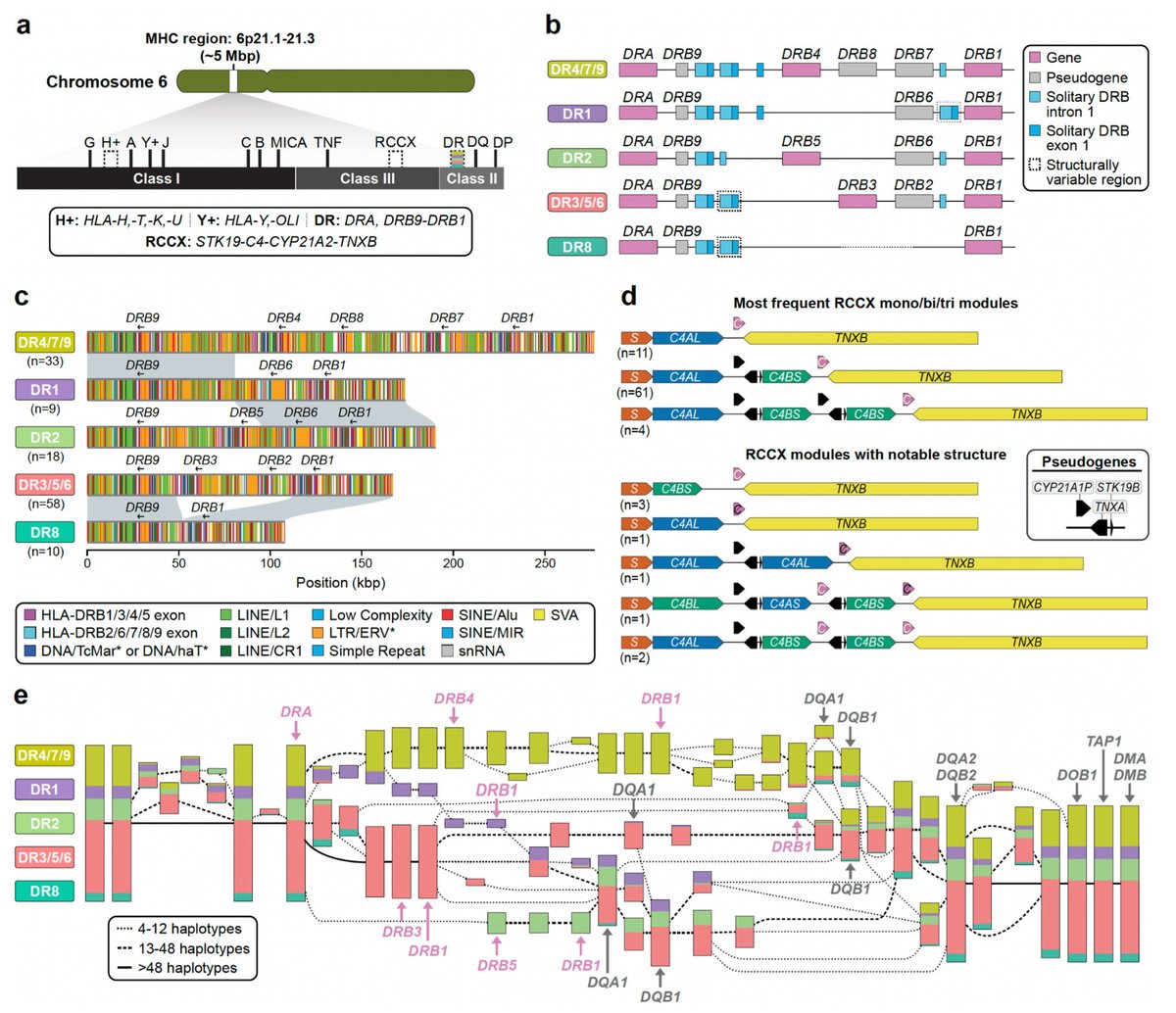
RT @lh3lh3: Want to annotate HLA/KIR genes and genotype them in a phased assembly? Try Immuannot, a new method from our group. It can ident….
0
64
0
RT @usadellab: A long journey driven by @tobiasmarschal in collaboration with Gunnar and I from @cepas_1 and @rfinkers. Rebecca was relentl….
link.springer.com
Potato is one of the world’s major staple crops, and like many important crop plants, it has a polyploid genome. Polyploid haplotype assembly poses a major computational challenge. We introduce a...
0
7
0
RT @ReichertLab: Leiter/Leiterin für das Genomics und Transkripromics Labor am BMFZ in Düsseldorf gesucht! . Wunderbare Gelegenheit mit Sta….
0
9
0
RT @WGGC_de: -NGS webinar series-.9th November Thursday 11:00 CEST. online-meet Philipp Schiffer - BioC2 priming biodiversity genomics in….
0
3
0
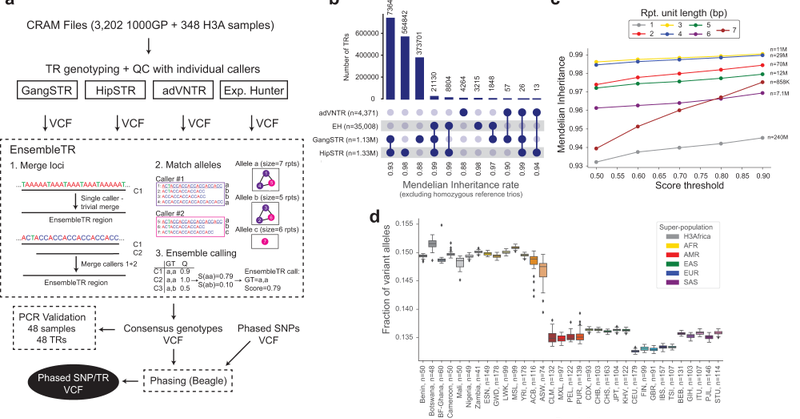
RT @HeliaZJ: Now out in @NatureComms! We developed EnsembleTR, an ensemble method to combine genotypes from 4 major tandem repeat callers a….
nature.com
Nature Communications - Tandem repeats (TRs) comprise some of the most polymorphic regions of the human genome but are difficult to study. Here, the authors develop an ensemble-based genotyping...
0
10
0
RT @AlexDilthey: We are hiring: Do you want to contribute to preventing the spread of multi-resistant bacteria by building browser-bashed d….
0
10
0
RT @pille_hallast: I am thrilled to share our study on the de novo assembly and characterisation of 43 diverse human Y chromosomes, publish….
0
49
0
RT @WGGC_de: Our Bio-Data Science Evenings in Düsseldorf are back, where we meet biodata enthusiasts on-site & have a great time together!….
0
9
0
RT @yafmao92: 🎉 Exciting news! Our lab's first publication in Genome Biology is out now! We've delved into the large-scale genomic differen….
0
26
0
RT @dkoppstein: Two #opportunities for bioinformatician and HPC team lead with Peter Ebert and @tobiasmarschal at @UniklinikDUS! Work on hu….
0
4
0