Tobias Fehlmann
@tobias_fehlmann
Followers
100
Following
1K
Media
1
Statuses
900
Joined September 2016
RT @niklaskemp: Happy to share CREsted, our latest package on enhancer modeling! CREsted allows for training sequence-based deep learning m….
0
42
0
RT @davidjglassMD: RNAseq is commonly performed. We compared N=30 wild-type Bl6 mice to an N=30 heterozygotic knockout with a phenotype, a….
biorxiv.org
Determining the appropriate sample size (N) for comparative biological experiments is critical for obtaining reliable results. In order to determine the N, the usual approach is to perform a power...
0
217
0
RT @juliettevdnb: Excited to launch 'Biotech Chronicles', a podcast exploring the future of biotech. Kicked off….
open.spotify.com
Podcast · Juliette Vandenbroucque · Welcome to Biotech Chronicles, a forward-looking podcast on how biotech is transforming the future of humanity and healthcare. In each episode, we will dive into...
0
4
0
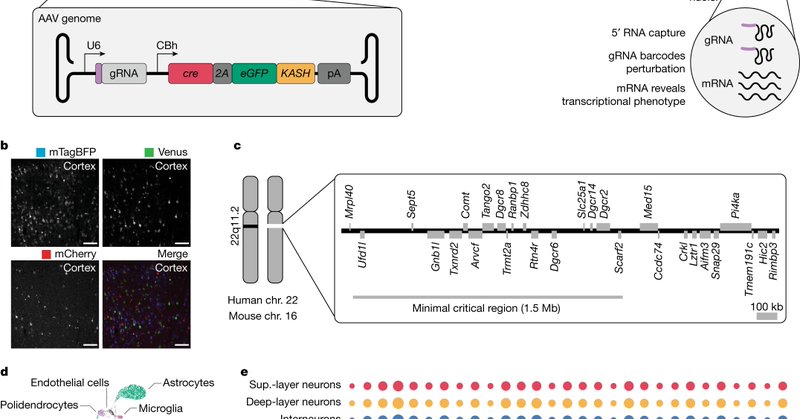
RT @AntonioSantinha: 🚨🚨🚨.Ever wanted to do in vivo single-cell CRISPR screens but don’t know where to start? Then check our latest publicat….
nature.com
Nature - An in vivo single-cell CRISPR screening method identifies transcriptional phenotypes of 22q11.2 deletion syndrome associated with a broad dysregulation of a class of disease susceptibility...
0
68
0
RT @soumithchintala: No More GIL!.the Python team has officially accepted the proposal. Congrats @colesbury on his multi-year brilliant ef….
discuss.python.org
Posting for the whole Steering Council, on the subject of @colesbury’s PEP 703 (Making the Global Interpreter Lock Optional in CPython). Thank you, everyone, for responding to the poll on the no-GIL...
0
1K
0
RT @fchollet: We're launching Keras Core, a new library that brings the Keras API to JAX and PyTorch in addition to TensorFlow. It enables….
0
784
0
RT @schwabpa: Does form follow function in cells?. We recently explored the use of machine learning to associate cell shape with multi-omic….
0
42
0
RT @dvbro0: Happy to present our new preprint! We ran a head-to-head comparison of sample multiplexing reagents for single-cell RNA-Seq on….
biorxiv.org
Single-cell RNA sequencing (scRNA-Seq) has emerged as a powerful tool for understanding cellular heterogeneity and function. However the choice of sample multiplexing reagents can impact data quality...
0
33
0
RT @gama_search: Our paper with @jeanphi_vert and @VallotCeline on how best to analyse single-cell histone modifications is finally out on….
genomebiology.biomedcentral.com
Background Single-cell histone post translational modification (scHPTM) assays such as scCUT&Tag or scChIP-seq allow single-cell mapping of diverse epigenomic landscapes within complex tissues and...
0
16
0
RT @fcrameri: Scientific colour maps 8.0. #useBatlow.#ScientificVisualisation #visualisation #Science #ColourPalet….
0
456
0
RT @frankstefansch1: Is Adam the best optimizer to train neural networks?🤔.We don't know. And we won't know until we test training algorith….
0
115
0
RT @JCoolScience: Big News🚨: #CZCellxGene Discover Census launched today! Built from >500 datasets, Census gives y….
0
154
0
RT @sama: i think @bettslacroix is the best biotech ceo i've met, and quietly leading a revolution that will result in us living healthy lo….
retro.bio
Our mission is to extend healthy human lifespan by 10 years.
0
91
0
RT @Oliver__Hahn: Some may have heard the news that I'll be soon joining Calico (more about that soon) .I'm looking for a (senior) research….
calicolabs.com
0
13
0
RT @sinabooeshaghi: Excited to share a new preprint with @XiChenUoM and @lpachter introducing seqspec for describing, organizing, and annot….
0
66
0
RT @steman_research: 🧬💻🔥 We are excited to launch #CuratedAtlasQueryR! millions of cells at your fingertips. A thread 🧵. The harmonized a….
0
137
0
RT @fabian_theis: Excited to finally see ExpiMap out @NatureCellBio - led by @Mohlotf & Sergei Rybakov, we inform single-cell embeddings by….
0
44
0
RT @junyue_cao: Excited to share our latest preprint from Cao Lab - PerturbSci-Kinetics, led by brilliant @xu_zxu! The technique captures s….
0
42
0