Rushin Gindra
@rushin_gindra
Followers
133
Following
610
Media
2
Statuses
55
PhD candidate @SaurLab at TranslaTUM and @helmholtz_ai, developing deep learning methods for Spatial Transcriptomics
Munich, Germany
Joined January 2017
RT @sophiajwagner: Excited to share TITAN, a multimodal foundation model for pathology trained on >330k WSIs, synthetic captions, and repor….
huggingface.co
0
9
0
It’s a struggle to know what are cells and whether us as humans can delineate categories of them. This article explained it so well! Worth a read. And if you’re interested to know more about my research I‘d be glad to explain over a cup of Coffee! ☕️😉.
nature.com
Nature - Scientists have more information than ever on how cells differ — but they still resist easy grouping.
0
0
1
RT @TranManuel: I am thrilled to announce HistoGPT, a vision language foundation model that generates highly accurate pathology reports fro….
0
44
0
RT @LukasHeumos: Our paper presenting @scverse_team - our multi-institution open-source software project to address storage and analysis ne….
0
36
0
RT @fabian_theis: Glad to see our review on best practices for single-cell analysis across modalities out @NatureRevGenet! In a big team ef….
0
223
0
RT @JCashaback: Good luck to all the PhD students this week, where all their family try to understand what they do and ask when they will b….
0
1K
0
Can't agree more! Also, learning about biology "empirically" can be difficult + not reflect the real world scenario. We should focus more on interpretability and robustness!.
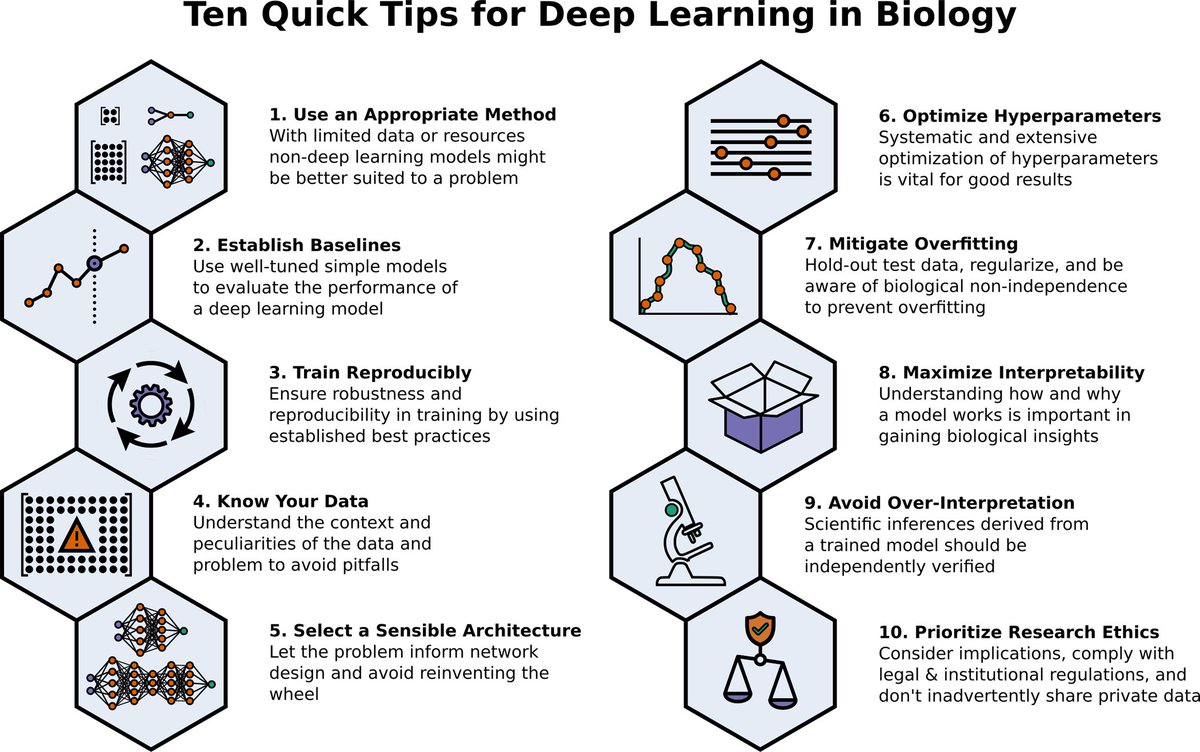
Nice 10-tips-article by @Benjamin_D_Lee et al @PLOSCompBiol on deep learning in biology. I like the opening of the box focus to „maximize interpretability“ - ideally we not only predict but want to learn about biology from our models.
0
0
2
Now that we have a pretty cool documentation for sc data analysis best practices, it's time we step up the game and develop spatial data analysis tools. .
1/n Single-cell data analysis is complex with many tools to choose from - but which ones work best and should be used when? Led by @lukasheumos and @AnnaCSchaar, we wrote a free online book that aims to guide single-cell data analysts:
0
0
1
RT @mertrory: Some advice I give my graduate students about how to keep on top of the literature in their field. A 🧵: 1/.
0
2K
0
RT @cs_sop_org: As PhD application deadlines approach, we are super excited to announce created by @zhaofeng_wu @a….
0
203
0
RT @Aiims1742: Another glorious preprint from the @hopkinskimmel Convergence team (Dr. @ltkagohara et al): .Spatial transcriptomics of FFPE….
0
34
0
As a computational researcher, I am curious to know how much of a variation an expert pathologist can observe in the cellular morphology with changes in it's genomic content? Is that morphological variation measurable?.@SaurLab would you like to share your thoughts?.
0
0
4
RT @jbhuang0604: How to develop a productive routine?. When transiting from structured learning environments (e.g., courses) to unstructure….
0
43
0
Looking forward to connecting with a few smart researchers and understand the interesting projects using AI at #HelmholtzAIcon2022.
@fabian_theis is introducing @helmholtz_ai at the Helmholtz Conference 2022 in Dresden and checking „state of you“ #helmholtzAICon
0
0
0
RT @scverse_team: We are very excited to announce scverse (, a new consortium around the core Python packages for s….
scverse.org
Foundational tools for single-cell omics data analysis
0
226
0