Sophia J. Wagner
@sophiajwagner
Followers
305
Following
250
Media
8
Statuses
34
PhD student @helmholtz_ai and @TU_Muenchen. AI for Computational Pathology
Joined April 2021
RT @ValentinKoch_: 🧬 Histopathology just got smarter! CellPilot is an open-source framework for cell & gland segmentation, trained on 675,0….
0
3
0
Excited to share TITAN, a multimodal foundation model for pathology trained on >330k WSIs, synthetic captions, and reports🔬⚡.co-lead w @TongDing99 @GreatAndrew90 @richardjchen.Thanks to @AI4Pathology @p_tingying!. Preprint: Model:
huggingface.co
⚡️📣After the success of our previous pathology foundation models UNI ( and CONCH (, we are now announcing TITAN (, a new state-of-the-art whole slide level foundation model trained on >330k pathology slides
1
9
34
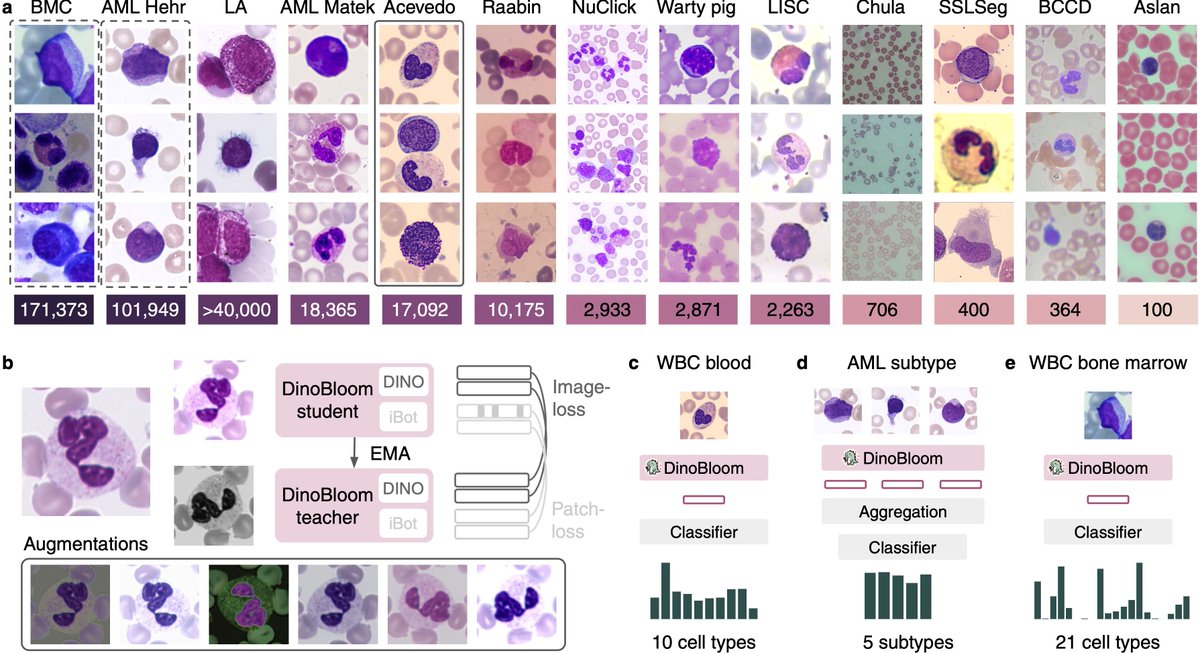
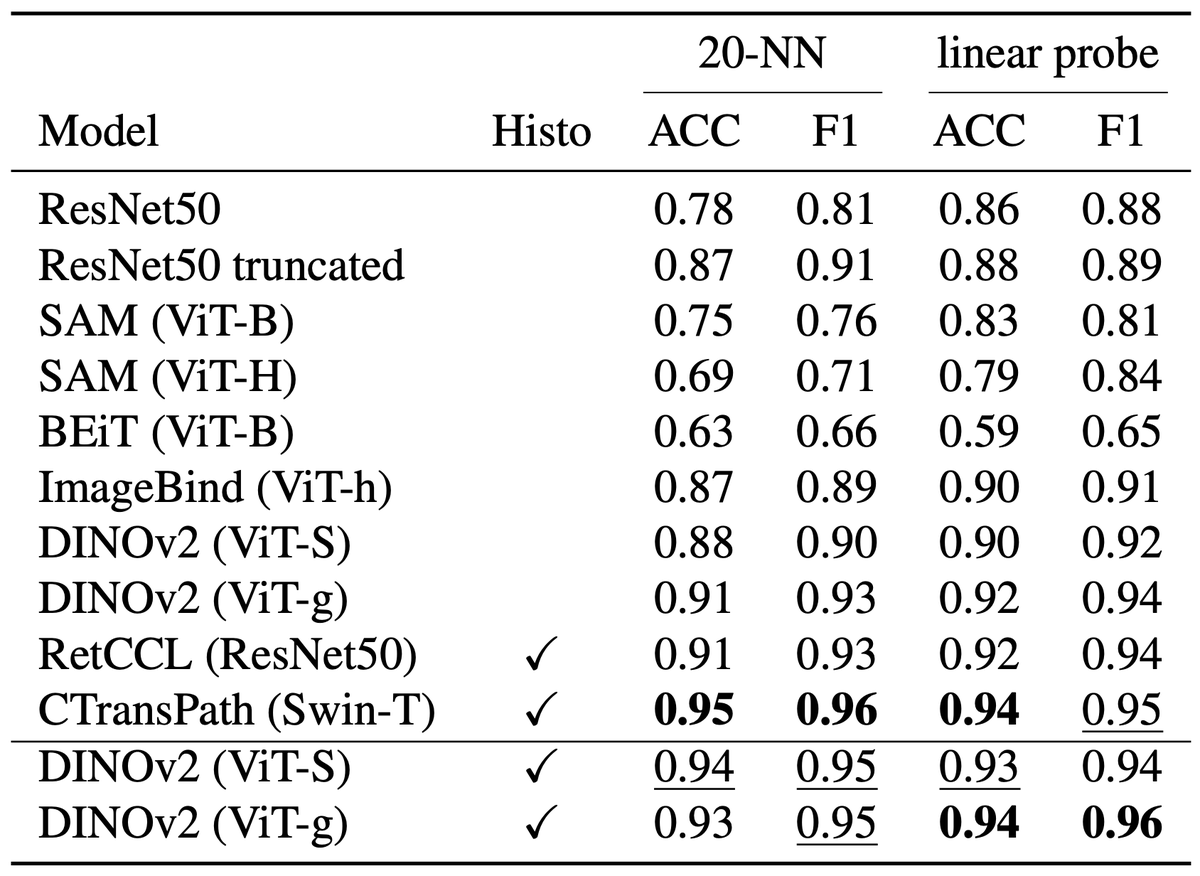
Join us at our poster T64 at #MICCAI2024 to talk about DinoBloom 🦖🌺 the first feature extractor in hematology for bloodsmears🩸.All models are available at .Developed w @ValentinKoch_, supervised by @p_tingying @MarrCarsten @HelmholtzMunich @TU_Muenchen
0
3
26
RT @ValentinKoch_: 1/ 🧵Introducing UNICORN,🦄 the first #AI model for integrating multi-stain #histopathology images for classification, des….
0
7
0
RT @AI4Pathology: ⚡️📣Delighted to announce MMP, a prototype-based multimodal framework combining histology and transcriptomics for cancer o….
0
39
0
RT @AI4Pathology: What would you do with 1000+ spatial transcriptomics samples with corresponding H&E-stained whole-slide images? Meet HEST….
0
104
0
RT @ValentinKoch_: Join us tomorrow at 9:45 at #ISBI Lecture hall if you want to get insights on how to train your custom #histopathology f….
0
3
0
RT @GreatAndrew90: I am tremendously excited to announce that my work on 3D computational pathology has been published in @CellCellPress !!….
0
9
0
Happy to share DinoBloom 🦖🌺 for generalizable cell embeddings of blood🩸and bone marrow💉smears in #hematology developed w @ValentinKoch_, take a look at this thread👀.preprint: github: @HelmholtzMunich @MarrCarsten @p_tingying.
github.com
Blood Cell Foundation Model based on DINOv2. Contribute to marrlab/DinoBloom development by creating an account on GitHub.
Excited to introduce DinoBloom🦖🌺, an open-source foundation model family (Vit-S to G) for general cell embeddings in #hematology🩸 Trained w mod. #DINOv2 on 13 datasets w 380,000+ WBC images of blood & bonemarrow. Evaluated on AML subtype & cell classification (knn, lin probe)
0
0
25
thanks to our supervisors @ja_schnabel @p_tingying @MarrCarsten !!.the work was done at @CompHealthMuc @HelmholtzMunich and @TU_Muenchen .4/4.
0
1
6
⚡️Want to save compute when training your feature extractor for histopathology?🔬💻#CPath.Study on 1(!) GPU for 2 CRC tasks led by our fantastic master student @roth_benedikt13, co-supervised with @ValentinKoch_ .pre-print: code: 1/
3
6
22
Enjoyed presenting parts of our work on biomarker prediction on colorectal cancer at the #ICCV2023 workshop on automated medical diagnostics. Happy to catch up if you’re around :).@ICCVConference @p_tingying @jnkath
2
3
49
RT @ValentinKoch_: 🚀 FIRST-EVER Marr & Peng Lab Hackathon in beautiful Oberammergau! 👩💻 Was a great time to kickstart some promising proje….
0
3
0
RT @HelmholtzMunich: 📝Hope for Colorectal #Cancer.Using #AI, scientists from #HelmholtzMunich & TU Dresden boost #precision #medicine in #o….
0
11
0
Thank you so much @jnkath for the opportunity to collaborate with you and all the amazing researchers involved☺️I had a great time in your lab last summer!.
0
0
4
Excited to share our new approach for “Transformer-based biomarker prediction from colorectal cancer histology”, just published in @Cancer_Cell @p_tingying @ja_schnabel @HelmholtzMunich @TU_Muenchen @EKFZdigital.
cell.com
Wagner et al. show that transformer-based prediction of biomarkers from histology substantially improves the performance, generalizability, data efficiency, and interpretability as compared with...
Excellent result of @sophiajwagner‘s 2022 summer internship in our group 🤩🤩 just out in @Cancer_Cell: Transformer-based biomarker prediction from colorectal cancer histology @EKFZdigital @DKFZ @tudresden_de @NCT_UCC_DD @NCT_HD @HelmholtzMunich.
2
3
31
RT @helmholtz_ai: Great success at our parallel Session 2b: "Applied AI" today! Thanks to our outstanding speakers Annika Bande, Yunfei Hua….
0
4
0