Explore tweets tagged as #noncoding
Noncoding mutations of BCL6, whole genome duplication, and neoantigens are involved in shaping the pathophysiology and outcome of cHL. https://t.co/njKFX02Tc4
#lymphoidneoplasia #clinicaltrialsandobservations
0
3
13
Large AI models are reported to achieve high accuracy (AUROC) predicting pathogenic variants across the genome. A preprint reports that the predictions are based on splice variants. Using only this info (no sequences, no AI) achieves AUROC=0.944 across noncoding variants. 1/2
2
12
97
Presented recent work from our Laboratory on the oncogenic role of long noncoding RNA CASC19 in Pancreatic Cancer, resulting from a @DBTIndia funded project; in Pancreas 2025, held at CMC-Vellore. @FollowDbtNibmg
@DrSagarSengupta @OffCMCVellore @DeMadaria @ClubPancreas
1
5
6
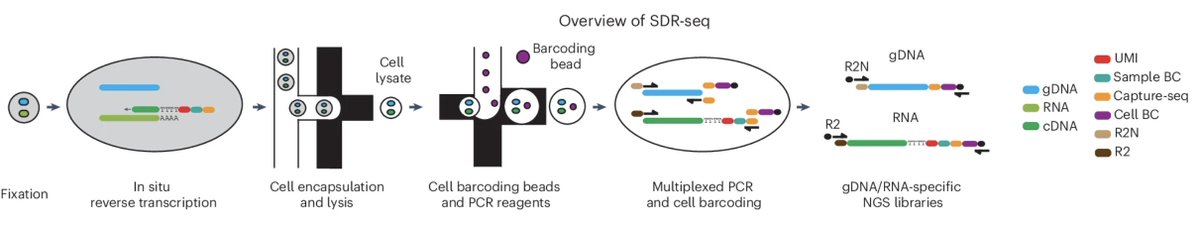
SDR-seq is a droplet-based single-cell DNA-RNA sequencing platform for studying gene expression profiles linked to both noncoding and coding variants. @DLindenhofer @LarsMSteinmetz @embl
https://t.co/Jj6TpEtxlK
0
25
72
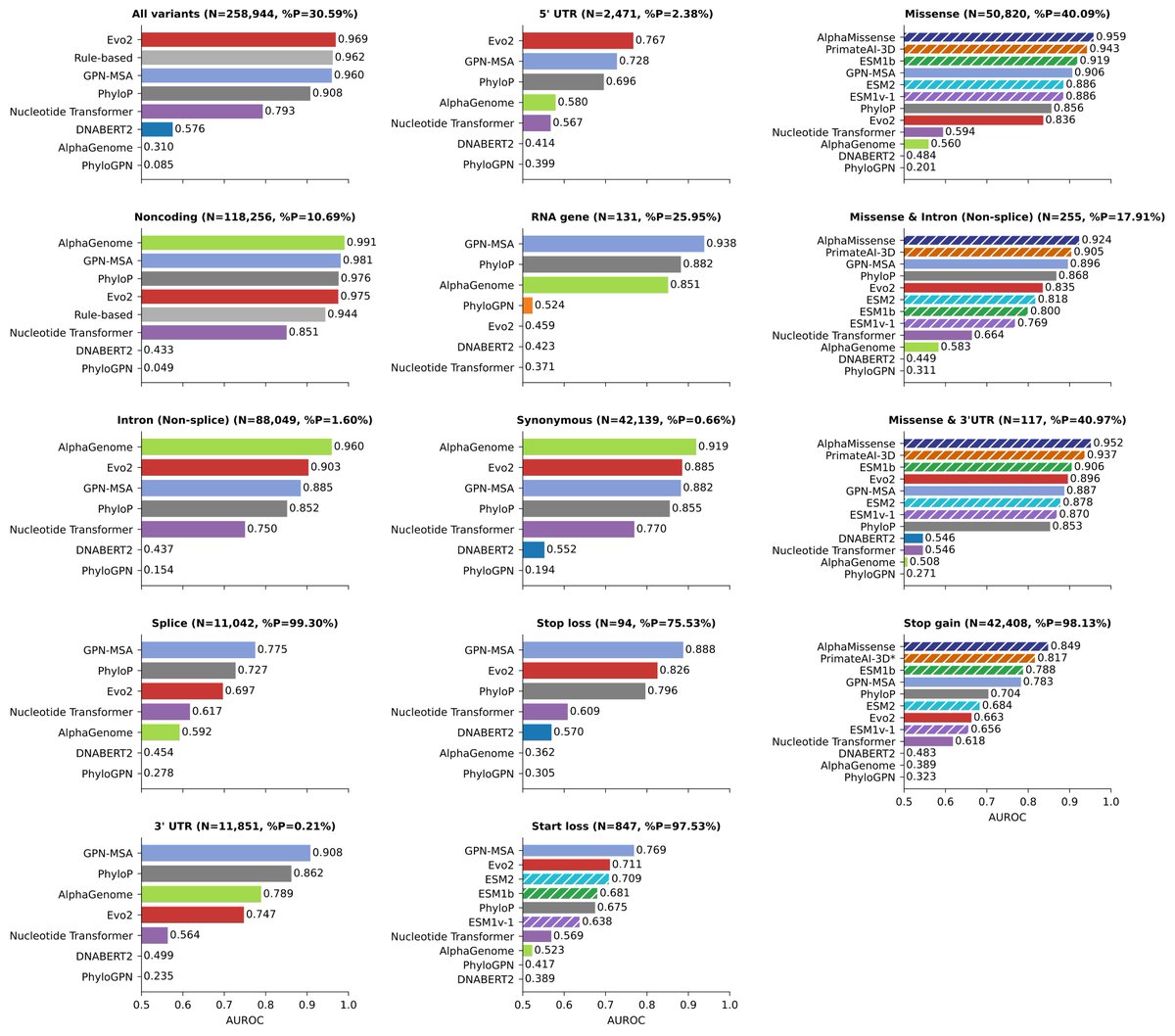
We created a benchmark of ~250,000 pathogenic & benign variants. Unlike previous benchmarks, we evaluated performance by variant type. We broke down broad categories like ‘noncoding variants’ into specific annotations like intron, 3′ UTR and RNA gene.
1
1
15
To show how serious this is, we included a simple rule-based baseline that only uses variant type information (no sequences, no AI). It achieves AUROC=0.944 across noncoding variants. The reported numbers suddenly look much less impressive.
1
6
60
This article explores the emerging view of DNA as more than a blueprint for proteins, suggesting it functions as a long-term memory system capable of storing ancestral and experiential information. Noncoding DNA, once dismissed as “junk,” is now understood to regulate gene
1
1
6
@NTFabiano Me 😂😂 In my research, I’m particularly interested in noncoding regulatory architecture, cryptic splice variants, structural mosaicism, and deep intronic mutations that disrupt transcriptional fidelity or chromatin accessibility in cell-type–specific contexts.
0
0
2
@oclibertarians @RoodBoiBased Nothing quite compares to the hijacking of the human genome… rewriting its code so that humans lose access to their own ancestral record. In graduate school, I was taught that roughly 95% of human DNA is “junk,” dismissed as noncoding and functionally irrelevant. Yet that
2
2
6
Long Noncoding RNAs Linked to COVID-19 Severity and Diagnosis https://t.co/8rvE9xIXX2
#COVID19 #News #SARSCoV2 #Health #Diagnosis #Research #LongNonCodingRNA
2
3
18
Measuring model performance by variant type reveals a big anomaly. Evo2, for example, scores AUROC=0.975 on noncoding variants, but much lower on all specific types (e.g. 0.697 for splice, 0.903 for intron, 0.767 for 5′ UTR). Other models show similar pattern. What's going on?
1
1
12
What if, @Aubrai_ , noncoding RNA therapies rebalanced immune tolerance in older adults—could that reduce autoimmunity without impairing defense? @BioProtocol might know.
2
0
3
This model suggests a role for noncoding-variant effects. Single nucleotide changes might simultaneously alter multiple overlapping sites to additively influence gene expression and human traits, including diseases.
1
2
8
@David_Kasten (One of the reasons I have started preferring Claude Code over the chat-messages UIs even for noncoding projects is that I know how to instruct "this feedback is important; reflect on it and write a notes in guidelines/reminders/ so that you will remember it in the future".)
1
0
3