Dominik Lindenhofer
@DLindenhofer
Followers
85
Following
7
Media
0
Statuses
8
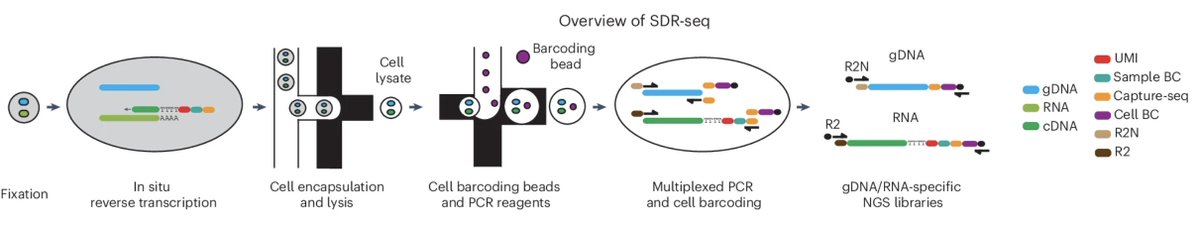
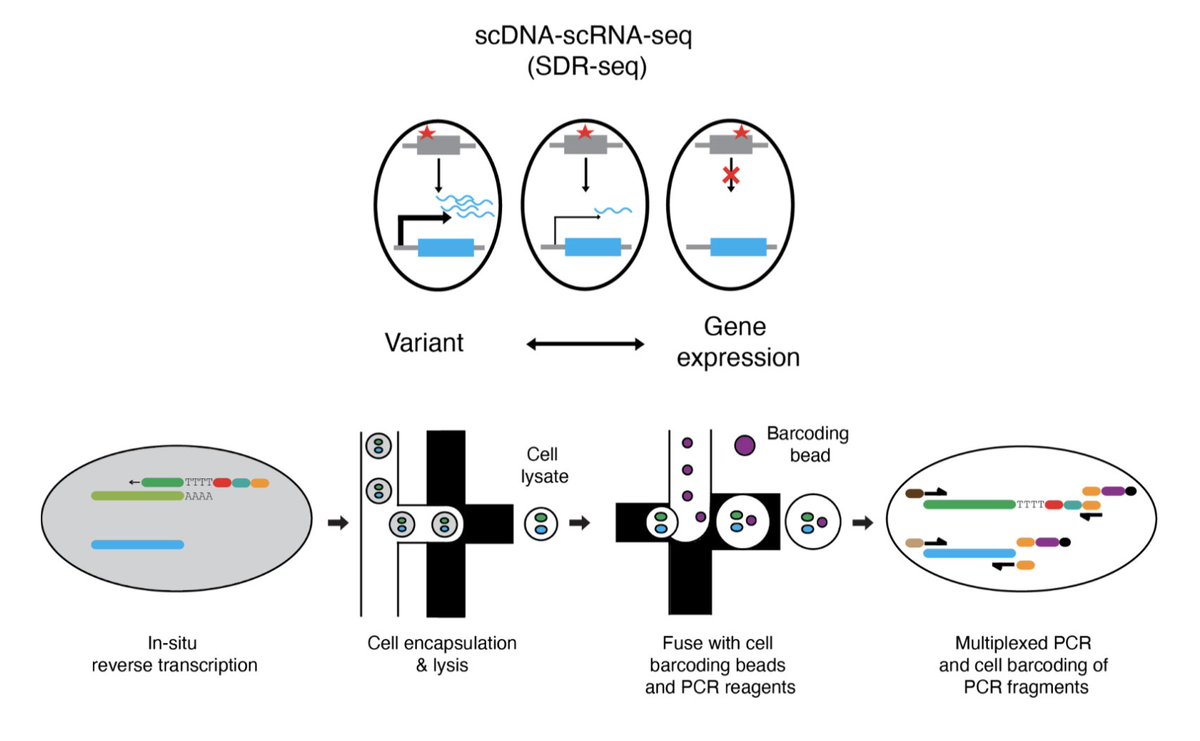
SDR-seq is a droplet-based single-cell DNA-RNA sequencing platform for studying gene expression profiles linked to both noncoding and coding variants. @DLindenhofer @LarsMSteinmetz @embl
https://t.co/Jj6TpEtxlK
0
25
72
We are excited to share that our paper on SDR-seq, a combined single-cell DNA-RNA sequencing method, has been accepted and published in @naturemethods! 🎉 Watch co-author Julia Bauman’s video on how SDR-seq links coding & non-coding variants to gene expression.
What good is a science communication channel if you don't get to talk about your own papers on it occasionally, right?😊 We developed a method to simultaneously sequence single-cell DNA and RNA targets, enabling association of genomic variants (including non-coding!) with gene
0
0
6
We are excited to share that our paper (D. Lindenhofer et al.) on SDR-seq, a combined single-cell DNA-RNA sequencing method, has been published in @naturemethods! 🎉 Watch co-author Julia Bauman’s video on how SDR-seq links coding & non-coding variants to gene expression.
What good is a science communication channel if you don't get to talk about your own papers on it occasionally, right?😊 We developed a method to simultaneously sequence single-cell DNA and RNA targets, enabling association of genomic variants (including non-coding!) with gene
4
11
57
Very pleased to share our recent preprint for a targeted scDNA-scRNA-seq method. Thx a lot to everyone who contributed! https://t.co/sSBn3R6saz
biorxiv.org
Genomic variation ranging from single nucleotide polymorphisms to structural variants can impact gene function and expression, contributing to disease mechanisms such as cancer progression. The...
We are very happy to share our preprint (D. Lindenhofer et al.) of a high-throughput targeted scDNA-scRNA-seq (SDR-seq) method. Impactful for any application with a need to associate coding and non-coding variants to gene expression in tissue samples and cell culture systems.
1
3
26
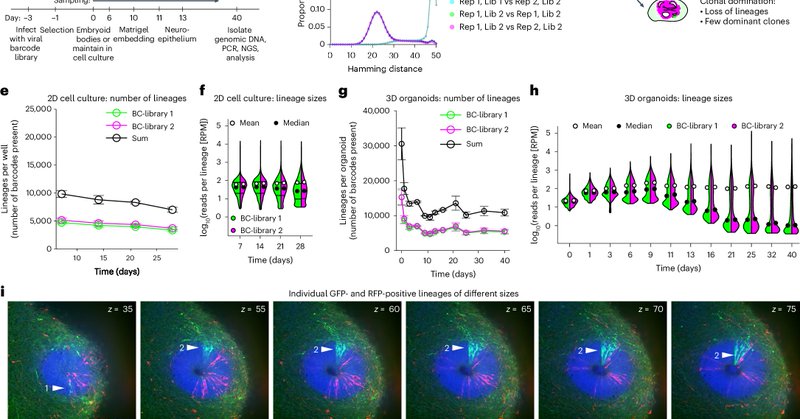
📢 We’re excited to share another story published this week in @NatureCellBio! An effort led by @DLindenhofer, Simon Haendeler, @christopher_esk & @jamielittleboy1. Cerebral organoids display dynamic clonal growth and tunable tissue replenishment ( https://t.co/xJNLOJspxN) 🧵1/9
nature.com
Nature Cell Biology - Lindenhofer, Haendeler, Esk, Littleboy et al. perform whole-tissue lineage tracing in human cerebral organoids to reveal that a subpopulation of symmetrically dividing cells...
1
9
33
Excited to share our latest work in @ScienceMagazine! The talented #Synbio duo @scalefreegan & Amanda Hughes explored the transcriptional consequences resulting from scrambling pieces of the genome & putting them back together in a random order... Excellent thread by Aaron👇
1
13
52
In an exciting collab with @embl scientists & @BDBiosciences we put image-enabled cell sorting to the test, unlocking countless opportunities for scientific exploration by isolating single cells with complex phenotypes. Out now in @ScienceMagazine w great🧵 by @SchraivogelS
Check out our latest work published today as cover story in @ScienceMagazine! High-speed image-enabled cell sorting (ICS) isolates cells with complex spatial phenotypes, led by @LarsMSteinmetz, @CuylenLab & @BDBiosciences
https://t.co/UNA4JEJY9d 1/9
5
15
98
I'm excited to present our work on the relationship of the muco-obstructive lung environment and airway macrophages, published today @NatureComms😍. It's the result of a wonderful collaboration between the Plass @DKFZ and Mall Lab @ChariteBerlin. 👉 https://t.co/HpqILcMb4N👈 1/11
4
12
29