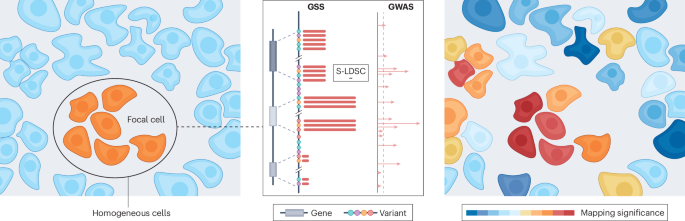
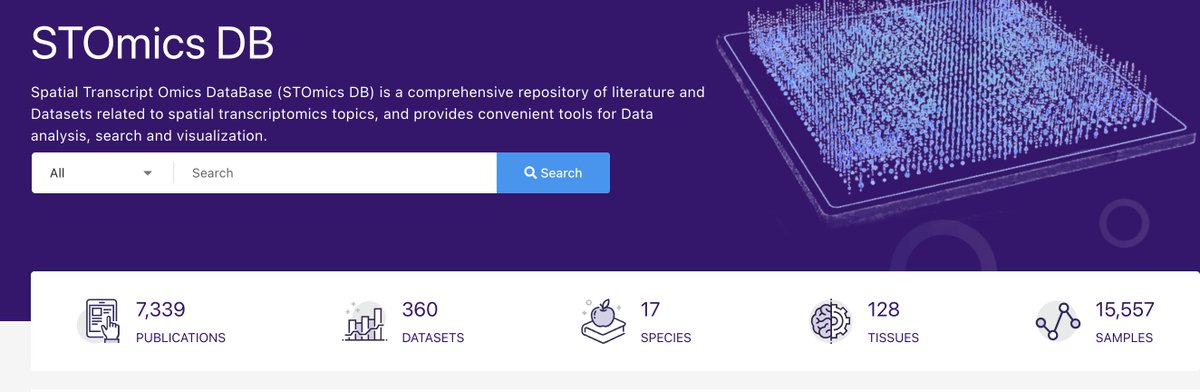
Explore tweets tagged as #Spatial_Transcriptomics
Thrilled to share our new paper on spatialMNN! . It's a fast, scalable, and accurate method to integrate multiple spatial transcriptomics (SRT) datasets and identify shared spatial domains. #SpatialTranscriptomics #Bioinformatics #Genomics #RStats
2
5
9
🚨 We’re hiring in Dr @Wi_Roman's Group. The Roman group study how cells communicate to keep organs healthy – and how this goes wrong in disease & ageing. Work with iPSCs, biomaterials, organ-on-chip & spatial transcriptomics to advance muscle biology & regenerative medicine.
0
3
8
Systematic evaluation of robustness to cell type mismatch of deconvolution methods for spatial transcriptomics data. #SpatialTranscriptomics #DeconvolutionMethods @biorxiv_bioinfo .
0
1
6
🌱 Xenium breaks barriers in plant spatial biology!. Thick cell walls, autofluorescence & complex tissues have long challenged spatial transcriptomics. A new bioRxiv preprint shows an optimised @10xGenomics Xenium workflow for Medicago truncatula roots & nodules.
1
2
3
Integration of spatial protein imaging and transcriptomics in the human kidney tracks the regenerative potential of proximal tubules | @ScienceAdvances
0
6
31
#UTSW team led by Prithvi Raj, Ph.D., now boasts innovative tools like NanoString’s CosMx and 10x Genomics’ spatial transcriptomics systems—enabling researchers to map gene expression at single-cell and spatial resolution. Learn more: #BiomedicalResearch.
0
0
7
In the Brain Resilience Lab, Shon has contributed to flagship projects that mapped the molecular landscape of human brain aging and led collaborations using next-generation sequencing, histology & spatial transcriptomics. He’s now starting his neuroscience PhD at @UCSF. Congrats!
1
2
9