Gervasio's Research Group (Protein Dynamics)
@gervasiolab
Followers
2K
Following
897
Media
45
Statuses
378
Prof F.L. Gervasio's lab at UCL and Uni Geneva. Follow us for the latest #compchem and, #DrugDiscovery updates from the lab. @gervasiolab.bsky.social
London, England
Joined March 2019
A structure-based drug design conference at an impressive venue but more so with fantastic friends @gervasiolab @AdrianMulholla1 @RommieAmaro Qiang Cui Matteo Dal Peraro
0
2
13
📷 How do proteins really fold? Our latest @JPhysChem Lett. paper unveils a new enhanced sampling MD strategy to investigate it in atomistic resolution by focusing on water and side-chain interactions. Check it out :
pubs.acs.org
Protein folding remains a formidable challenge despite significant advances, particularly in sequence-to-structure prediction. Accurately capturing thermodynamics and intermediates via simulations...
0
9
35
This paper didn’t go viral but it should have. A tiny AI model called HRM just beat Claude 3.5 and Gemini. It doesn’t even use tokens. They said it was just a research preview. But it might be the first real shot at AGI. Here’s what really happened and why OpenAI should be
347
1K
10K
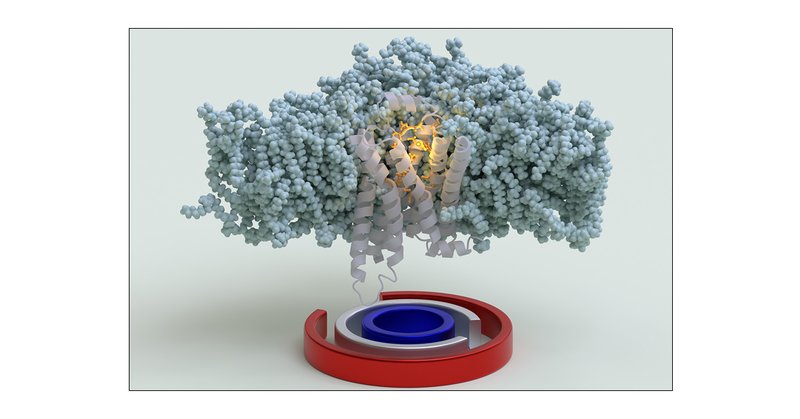
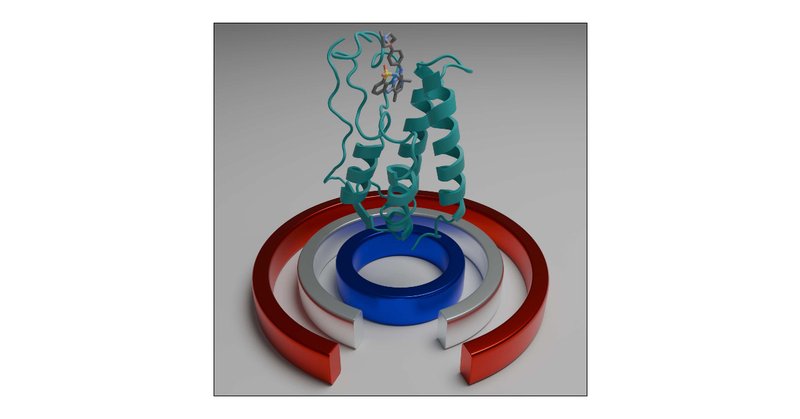
New in @JCIM_JCTC : We've mapped the complete activation pathway of the β1-adrenergic receptor using our OneOPES enhanced sampling method and tailored CVs. Our approach reveals how sodium ions, water networks & protein microswitches work together. https://t.co/UdpUqpziiF
#GPCR
pubs.acs.org
The beta-1 adrenergic receptor (ADRB1) is a critical target for cardiovascular drugs, yet our understanding of how it is activated remains incomplete. Capturing the concerted interplay of agonists,...
0
6
25
The corresponding News & Views from @gervasiolab, @TFroehlking and Simone Aureli is now live! https://t.co/jQ6Qae9f6b 🔓 https://t.co/m7LtYEbCWC
0
3
3
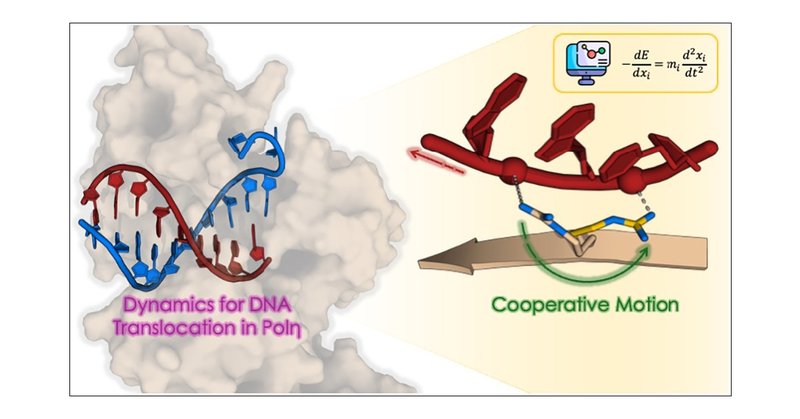
Our contribution in #JACS: deep-learning-guided simulations to detect the molecular mechanism for DNA translocation during nucleic acid polymerization. We loved it ! 😊 Congrats to co-authors! Take a look please ! In collab with @GroupParrinello
https://t.co/mSrza6Tfik
pubs.acs.org
The translocation of DNA in polymerase (Pol) enzymes is a critical step for Pol-mediated nucleic acid polymerization, essential for storing and transmitting genetic information in all living organi...
2
12
67
Happy New Year tweeps! A new review article in collaboration with Pande's lab at @JNJNews and @gervasiolab at @UNIGEnews on "Computational advances in discovering cryptic pockets for drug discovery" is now live @COSB_CRSB @ElsevierConnect
0
4
16
An initiative coordinated by Gareth Tribello, @BonomiMax, @BussiGio, Carlo Camilloni, and with fundamental contributions from our community. 𝕏 handles in 🧵:
1
2
10
🔴 LIVE in 25 minutes: Computational Protein Design Then & Now (1988-2024) A free one-hour webinar featuring: David Baker (UW) Steve Mayo (Caltech) Bill DeGrado (UCSF) Brian Kuhlman (UNC) https://t.co/OtdAq6HcPE
5
60
196
What is happening? HandMol is happening! Coming soon, multiuser, immersive, interactive simulations.
13
139
777
Valentin Miéville is one of the 2024 winners of the "#PhD Booster" programme launched by the Faculty of Medicine, which aims to promote young talent in #biomedical #research. His research "Small organs, great outcome: organoids against kidney cancer" https://t.co/1TE4jXyRTW
0
1
1
Much of the success of breakthroughs, such as AlphaFold, is thanks to a database of protein structures dreamed up in the 1960s by Helen Berman
nature.com
Nature - Pioneering crystallographer Helen Berman helped to set up the massive collection of protein structures that underpins the Nobel-prize-winning tool’s success.
17
274
880
I said that reaching Human-Level AI "will take several years if not a decade." Sam Altman says "several thousand days" which is at least 2000 days (6 years) or perhaps 3000 days (9 years). So we're not in disagreement. But I think the distribution has a long tail: it could take
I seriously wonder why Yann Lecun takes such different positions from Sam Altman and Dario Amodei. Sam and Dario have made it clear in their posts that they believe reasoning will be solved, if not now, then in a few years' time. What's more: in his last post, Dario said that AGI
163
164
2K
Congratulations to David Baker, Demis Hassabis and John Jumper!
BREAKING NEWS The Royal Swedish Academy of Sciences has decided to award the 2024 #NobelPrize in Chemistry with one half to David Baker “for computational protein design” and the other half jointly to Demis Hassabis and John M. Jumper “for protein structure prediction.”
0
0
13
On Friday I defended my PhD in computational drug discovery at @sciences_UNIGE It is based on these two publications: https://t.co/G9DHvaZT3F
https://t.co/Mjo6CLKTJH I want to thank my PI @gervasiolab and the members of the jury Bert de Groot and Jean-Luc Wolfender #compchem
0
1
5
Looking for an accurate and transferable strategy to calculate protein-ligand absolute binding free energies? Check out our latest paper now published in @JPhysChem with @M_Karrenbrock @ValerioRizzi33 @D_Lukauskis Simone Aureli @gervasiolab
https://t.co/jn66zS2I4m 1/🧵
pubs.acs.org
The calculation of absolute binding free energies (ABFEs) for protein–ligand systems has long been a challenge. Recently, refined force fields and algorithms have improved the quality of the ABFE...
1
10
43
Our commentary "A renewed call for open artificial intelligence in biomedicine" is now available as a preprint. We call for sharing training data, code, and model weights in biomedical artificial intelligence research. 1/
5
59
201
Excited to see our work on accelerating DeepLNE++ published in @JChemPhys at https://t.co/IxxjQLm0bH The updated link for tutorials: https://t.co/2IdfSI5c8d Many thanks to @ValerioRizzi33, Simone Aureli, and the entire @gervasiolab. Summary below: https://t.co/Ev5T0Tt1jF
github.com
DeepLNE speed-up via knowledge distillation. Contribute to ThorbenF/DeepLNE_PlusPlus development by creating an account on GitHub.
Checkout our #preprint of our latest manuscript on DeepLNE++, our new and improved deep-learning path-like collective variable https://t.co/BOL3QjfoQ6 by @TFroehlking, @ValerioRizzi33, Simone Aureli, @gervasiolab . Summary in thread:
0
4
10