Francesco Beghini
@chocophlan
Followers
366
Following
5K
Media
404
Statuses
6K
Postdoctoral Fellow at @Yale | Human microbiome | Bioinformatics | Go @Patriots!
Joined November 2009
A new study found that we share parts of our microbiome with people in our social networks, beyond family members. Dr. @NAChristakis joins us to discuss the research and how scientists can identify your friends—just by looking at your poop. https://t.co/CIZMzeirrr
1
6
7
This work was supported by the NOMIS foundation, with additional support via @SchmidtFutures @ericschmidt @PershingSqFdn @BillAckman @JMRothberg. Underlying cohort supported by @gatesfoundation & @NIH. New support for work on the biology of social life at #HNL via @paulg. 19/
2
2
38
News coverage of “Gut microbiome strain-sharing within isolated village social networks” via @nature is here: https://t.co/L5wYcnMZfd 17/
nature.com
Nature - Analysis of nearly 2,000 people living in remote villages in Honduras reveals who’s spreading gut microorganisms to whom.
1
10
89
It may prove to be the case that groups of inter-connected people might share phenotypes not only because of shared genes or transmitted behaviors, but also because of shared microbes. https://t.co/GWUvrPJUxa 16/
nature.com
Nature - An investigation into the relationship between network structure and gut microbiome composition among people living in 18 isolated Honduras villages reveals that strain-sharing can be...
2
6
71
This work has implications for a radical idea: diseases formerly thought to be biologically non-communicable (e.g., obesity, depression, hypertension, arthritis, etc.) may actually be (somewhat!) communicable, via the spread of the microbiome. https://t.co/GWUvrPJUxa 13/
nature.com
Nature - An investigation into the relationship between network structure and gut microbiome composition among people living in 18 isolated Honduras villages reveals that strain-sharing can be...
3
37
183
Clusters of microbiome species and strains occur within clusters of people in village social networks. It’s like clouds of microbes occurring within clusters of people. https://t.co/GWUvrPJUxa 11/
2
8
65
Among 301 people in 4 villages whose microbiome was re-measured 2 years later, we observe greater convergence in gut microbiome strain-sharing in connected versus otherwise similar unconnected co-villagers. 10/
1
2
49
We also observe that socially central people are more microbially similar to the overall village than socially peripheral people. But popular people resemble the microbiomes of individual friends less. 9/
1
4
65
Furthermore, gut microbiome strain-sharing extends to one's friends’ friends within social networks, indicating the relevance of a person’s broader social network, at two degrees of separation. 8/
1
6
56
We can actually predict who your friends are based on whether you have similar bacteria in your poop! And that metric outperforms other more usual social features, such as how you resemble your friends on a host of traits like age, sex, wealth, etc. 7/
4
51
212
Using both species-level and strain-level data, we show that microbial sharing occurs in many relationship types, notably including non-familial and non-household connections. 6/
2
6
68
"Gut microbiome strain-sharing within isolated village social networks" https://t.co/GWUvrPJUxa with @chocophlan, @JacksonPullman1, @mqdicer, @ShivkumarVs, @DrewPrinster, @adarshsingh110, RM Juárez, @eairoldi, @ilanabrito123 #HNL 2/
4
29
159
One more time for the people in the back: AI is a software wrapper around machine learning, machine learning is fancy words for statistics, thanks for coming to my TED talk
28
121
906
Environmental, socioeconomic, and health factors associated with gut microbiome species and strains in isolated Honduras villages
cell.com
The complex relationship between the human gut microbiome and host factors remains obscure. Shridhar et al. implement a population-wide phenotype association analysis in rural Honduras and quantify...
0
6
20
Environmental, socioeconomic, and health factors are associated with gut microbiome species and strains in isolated Honduras villages. New #HNL work with @ShivkumarVs, @chocophlan @mqdicer A Singh, R Juarez, & @ilanabrito123
https://t.co/aosdjmq31V
#microbiome #LMICs
1
2
4
Deeply sequenced microbiome data from non-industrialized settings are uncommon. In new #HNL work, we use metagenomic data from 1,871 people in 19 isolated Honduras villages to report associations between bacterial species and human phenotypes and factors. https://t.co/aosdjmq31V
2
5
19
Beside analysing nappies our mission is to share the joy of microbiome science by surprising our little helpers (nappy donors) w/GIANT microbes Let us introduce our plushies 🔵Candidatus Cibionibacter quicibialis 🟡Catenibacterium tridentinum ©UniTrento Ph.Federico Nardelli 3/3
10
6
53
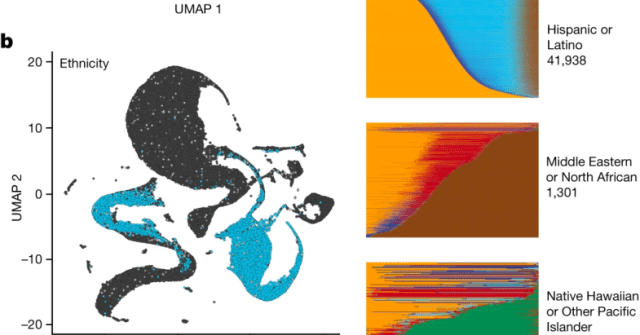
I wrote a blog post about scientific racism and the @AllofUsResearch figure illustrating their ancestry analysis. The post was inspired by a presentation of @DelaneyKSull at our lab DEI meeting organized by @NikhilaSwarna last week. https://t.co/fl6perfTGk cc @AllofUsCEO
liorpachter.wordpress.com
The All of Us Research Program, whose mission is to “to accelerate health research and medical breakthroughs, enabling individualized prevention, treatment, and care for all of us”, rec…
2
36
134
heard UMAP was popular on twitter, anyone have any advice on how to tighten up the clusters?
54
287
3K