Brian Hie
@BrianHie
Followers
10K
Following
3K
Media
98
Statuses
2K
AI for biology @Stanford and @arcinstitute
San Francisco
Joined October 2011
Today in @Nature, in work led by @aditimerch, we report the ability to prompt Evo to generate functional de novo genes. You shall know a gene by the company it keeps! 1/n
7
103
544
Nature research paper: Semantic design of functional de novo genes from a genomic language model https://t.co/vB53a2eIGM
nature.com
Nature - By learning a semantics of gene function based on genomic context, the genomic language model Evo autocompletes DNA prompts to generate novel genes encoding protein and RNA molecules with...
2
14
45
Excited to present the first major work after starting our lab at Stanford and the Arc this year: CRISPR-All, a unified genetic perturbation language for programming any major type of genetic perturbation simultaneously, in any combination, at genome scale, in human cells.
9
101
394
1/ Housing is the defining issue of our time. As California's average home prices approach $1 million, @CSElmendorf and Ed Glaeser, America's leading urban economists, call on California to stop the vetocracy and BUILD. MORE. HOMES. 🇺🇸 From today's @latimes:
162
199
2K
Excited to introduce CRISPR-All! – a flexible, high-throughput framework that combines virtually any perturbation modality (CRISPRko, CRISPRi, CRISPRa, shRNA, knock-in, base/prime editing, etc.) in the same cell, at scale. Paired with single-cell RNA-seq @10xGenomics and rich
Excited to present the first major work after starting our lab at Stanford and the Arc this year: CRISPR-All, a unified genetic perturbation language for programming any major type of genetic perturbation simultaneously, in any combination, at genome scale, in human cells.
5
85
408
Congratulations to Arc Science Fellow, @UcheMedoh, on being named a winner of the 2025 @ScienceMagazine & @SciLifeLab Prize for Young Scientists! His work solves a decades-old mystery in cell biology, revealing the enzyme responsible for BMP synthesis: https://t.co/e84WTyIKnE
2
12
128
How can we integrate genomic sequences and 3D chromatin structure to better understand cell-type specific genome organization?@biorxivpreprint @UW "Evo2HiC: a multimodal foundation model for integrative analysis of genome sequence and architecture" • Genomic sequences shape
1
10
43
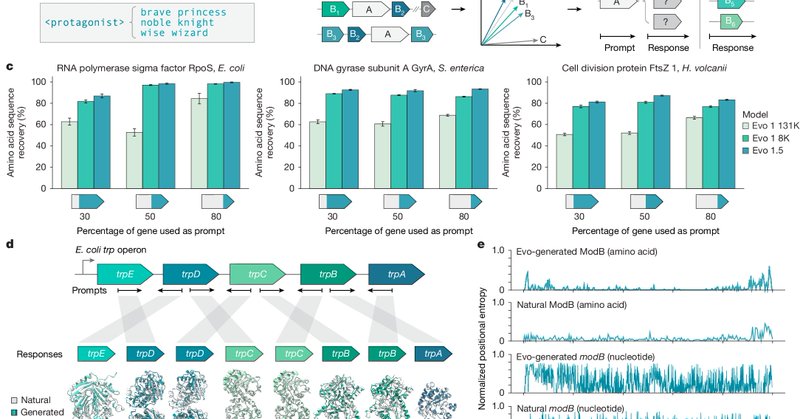
Published today in @Nature, @aditimerch & researchers from the @BrianHie lab report that the large-scale genomic model, Evo, is capable of using surrounding genomic context to produce novel, functional genes, enabling an emergent approach they've termed 'semantic design'.
4
25
91
What if we could autocomplete DNA based on function? Today in @Nature, we share semantic design—a strategy for function-guided design with genomic language models that leverages genomic context to create de novo genes with desired functions.🧵 https://t.co/P5qVJB3qIY
16
154
590
The ability to generate completely sequence-novel proteins provides an existence proof that -- with the right data and if prompted in the right way -- these models can generalize in very interesting ways. How do we elicit even more control over these models? n/n
1
0
24
There has been much progress in these genomic language models in a short amount of time, including the expansion of the models to eukaryotic genomes with Evo 2. What would the equivalent of semantic design look like in these organisms? 3/n
2
1
9
The details of the paper are in this excellent thread by @aditimerch: https://t.co/MUDWbLkcDa Some of my random thoughts are also provided below. 2/n
What if we could autocomplete DNA based on function? Today in @Nature, we share semantic design—a strategy for function-guided design with genomic language models that leverages genomic context to create de novo genes with desired functions.🧵 https://t.co/P5qVJB3qIY
2
1
16
Very exciting work demonstrating the emergence of in-context learning in Evo 2 on purely synthetic tasks.
For years since the GPT-2 paper, emergent in-context learning (ICL) from 'next-token' training has been treated as something deeply tied to 𝐡𝐮𝐦𝐚𝐧 𝐥𝐚𝐧𝐠𝐮𝐚𝐠𝐞. But … is it? Thrilled to share our latest result: 𝗚𝗲𝗻𝗼𝗺𝗶𝗰🧬 𝗺𝗼𝗱𝗲𝗹𝘀 𝘁𝗿𝗮𝗶𝗻𝗲𝗱 𝙤𝙣𝙡𝙮 𝗼𝗻
2
13
99
Closing the AI-to-lab loop is hard, especially if you want to test your WHOLE GENOME generator.. Viruses are the only genomes cheap enough to print en mass, but raise biosafety flags So @ArcInstitute chose phages! We went deep w/ @samuelhking & @driscoll_cl on how they did it:
2
11
53
We have an incredibly exciting research roadmap within a burgeoning new field and in a well-resourced academic setting. We are excited about open science, supporting independent career advancement, and cultivating the joy of science. n/n https://t.co/TtiBxwNeTR
evodesign.org
Developing biological AI for human good at Stanford University
3
0
17
The second position is within the general areas of synthetic genomics and synthetic biology, and again at the intersection with generative AI. More details here: https://t.co/lers0uLCnl 3/n
1
1
16
The first position is within the general areas of protein biochemistry, engineering, and evolution, as well as their intersection with AI systems for generative biology. More details here: https://t.co/3BKtexMfQ3 2/n
1
3
16
We are actively recruiting for two positions at the interface between biology and generative design. Backgrounds of particular interest are in protein biochemistry/evolution and synthetic genomics/biology. Please consider joining us! 1/n
8
92
348
Thanks for having me, it was a ton of fun! Proud to be a part of the Stanford SB community 🧬
Samuel King, @BrianHie Lab discusses his research on generative design of novel bacteriophages with genome language models @bioe_stanford 🧫 🧬
0
2
30
We're excited to welcome 28 new AI2050 Fellows! This 4th cohort of researchers are pursuing projects that include building AI scientists, designing trustworthy models, and improving biological and medical research, among other areas. https://t.co/8oY7xdhxvF
6
29
184