Arka Ray
@arka_ray1
Followers
41
Following
139
Media
0
Statuses
18
Graduate Student at University of Florida | Structural Biologist | Biophysicist | Membrane Proteins
Gainesville, FL
Joined May 2023
Thrilled to share our most recent work on the mechanisms by which cholesterol interacts with GPCRs.A huge thank you to the entire team @thakurnaveen7, @nilootw and @MatthewEddyLab #gpcr #nmr #proteinlipidinteractions #nanodiscs #membrane #cholesterol
https://t.co/bc6DZcyfL8
0
3
8
Excited to share our latest preprint of using 19F solid state NMR to study conformational dynamics of GPCRs. A huge thank you to Beining Jin and @MatthewEddyLab for all the support. https://t.co/aGEeWcntHb
#NMR #Membranemimetic #liposome #ssNMR #GPCR
biorxiv.org
Endogenous phospholipids influence the conformational equilibria of G protein-coupled receptors, regulating their ability to bind drugs and form signaling complexes. However, most studies of GPCR-l...
1
2
13
Check out our most recent preprint using 19F solid-state NMR to study GPCR conformational equilibria and compare that with receptors in detergents and lipid nanodiscs: https://t.co/vWX5t4klMJ Congrats to @arka_ray1 and Beining from the group! #NMR #NMRchat
2
9
48
🔥RNA & lipids in action 🧬! Our latest study, featuring @t_czer_jea as lead author, reveals how lipid membranes affect the activity of catalytic RNAs. @BCUBE_TUDresden @James_Saenz
Effects of lipid membranes on RNA catalytic activity and stability https://t.co/hWSKN0QUTJ
#biorxiv_synbio
2
13
43
Our new study on molecular mechanisms underlying cancer-associated mutations in a G protein is now available online https://t.co/YUpzONjQ2C Congratulations to @hugogdtc and great students in his lab and mine. @UFHealthCancer #NMR #NMRchat
0
6
16
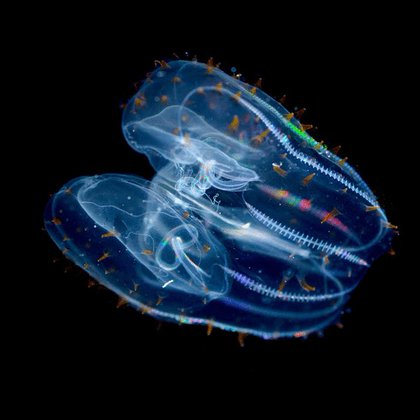
Today in @ScienceMagazine we report how lipids specialize for life under the crushing pressures of the deep sea and what this means more broadly for the composition of cell membranes (1/n)
science.org
Hydrostatic pressure increases with depth in the ocean, but little is known about the molecular bases of biological pressure tolerance. We describe a mode of pressure adaptation in comb jellies...
19
118
394
We are hiring an NIH-funded postdoc! Looking for a highly motivated researcher with a strong background in natural product isolation and structural characterization for #terpene NP discovery, #biosynthesis, #enzymology. Please RT! @UFChemistry
1
36
41
The critical importance of conditions: Reconciling GPCR functionality and biophysical findings
0
5
14
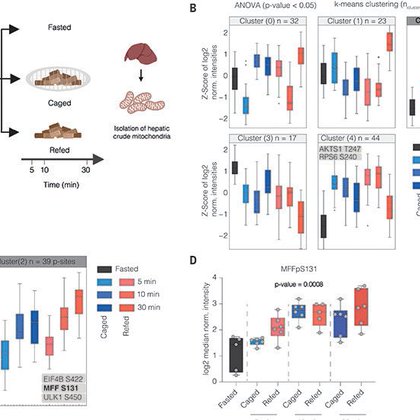
More than excited to share our latest study from my PhD work published in @ScienceMagazine! We demonstrate that food perception is sufficient to drive adaptational processes in hepatic mitochondria. For more details read the 🧵 and here: https://t.co/zbPjrGre3l Please RT.
science.org
Liver mitochondria play a central role in metabolic adaptations to changing nutritional states, yet their dynamic regulation upon anticipated changes in nutrient availability has remained unaddress...
11
44
186
Excited to share our latest pre-print! We map GPCR activation probability at the plasma membrane of live cells. Strikingly, we see ultra-long-lived (~5 min) nanodomains of high activation probability, while most receptors remain inactive. Interested? 🧵 https://t.co/9xp3ldFQ3z
4
21
68
Online now! Membrane mimetic-dependence of GPCR energy landscapes
0
9
41
Check out our latest publication that explores how membrane mimetics and their composition can affect GPCR energy landscapes. A huge thank you to @thakurnaveen7, our collaborators and @MatthewEddyLab for continued support and guidance.
0
2
25
Our new study comparing GPCR energy landscapes is out now in @Structure_CP with @thakurnaveen7, Ed Lyman, Ken Jacobson, and students from my group. If you work with nanodiscs or detergent micelles, you should check out this study. #NMR #NMRchat
https://t.co/MxAUq2Qxhw
1
22
77
A preprint of our new study on cancer-associated mutations in G proteins using biophysical and computational methods is available online. Thanks to @hugogdtc for a fruitful collaboration. #NMRchat #NMR #bioRxiv
https://t.co/nysQOjOFx1
1
8
26
Preprint of our latest work available on bioRxiv: "Membrane Mimetic-Dependence of GPCR Energy Landscapes" https://t.co/NP066saAMm NMR comparison of impact of membrane mimetics on GPCR activation. With @thakurnaveen7 @arka_ray1, Ken Jacobson, Ed Lyman & Zhanguo Gao. #NMR #NMRchat
biorxiv.org
Protein function strongly depends on temperature, which is related to temperature-dependent changes in the equilibria of protein conformational states. We leveraged variable-temperature 19F-NMR...
1
6
19
.@MatthewEddyLab @UFChemistry used 19F-NMR to study how cholesterol affects the conformational equilibria of the A2A adenosine receptor https://t.co/38lvYIavSJ
0
7
25