Andy Russell
@AndyRusss
Followers
881
Following
1K
Media
34
Statuses
730
Postdoctoral Fellow @broadinstitute @insitubiology (Chen Lab) @HSCRB . Ex: @SangerInstitute @Cambridge_Uni @UniofOxford. He/him IG: @ajcrscience
Cambridge, MA
Joined July 2009
Slide-tags adds spatial coordinates to single-cell sequencing measurements! Please explore the paper, published online today in @Nature! https://t.co/HIyNuhJyR4
nature.com
Nature - Slide-tags enables multiomic sequencing of single cells and their localization within tissues.
7
25
138
Excited to share a short article I wrote describing: Slide-tags - which enables spatial single-nucleus sequencing, allowing us to dive deeper into the complexity of tissue organization. Grateful for the amazing team and support from @insitubiology and @macosko! #ToolsOfTheTrade
Slide-tags enables spatial single-nucleus sequencing https://t.co/kqrJFQVQgY
#ToolsOfTheTrade by Andrew J. C. Russell @AndyRusss @broadinstitute @insitubiology @HSCRB
0
7
41
It was such a pleasure to meet everyone this past week in Heidelberg and be part of such a vibrant scientific community - thank you @EMBOFellows!
More than 100 @EMBOFellows are meeting in Heidelberg. The postdoctoral fellows present their research, attend a mentoring session, receive training on a variety of topics and more. #EMBOat60
0
0
7
Had fun talking about Slide-tags with @GenomeTDCC recently. Please check it out If you'd like an overview on what is is and what it can do! 🧬
Slide-tags: Combining single-nucleus genomics with spatial measurements 🧬🔬 Hear from @AndyRusss, @jacksonweir4, Naeem Nadaf, and @insitubiology as they describe the development and application of this impressive technology https://t.co/K6iYlKwuFy
0
3
10
We are happy to share our latest long-range, programmable mutagenesis toolbox - the Helicase-Assisted Continous Editing (HACE) system - to achieve target-specific mutagenesis of the endogenous genome. (1/11) https://t.co/boPlzlDBaE
biorxiv.org
A major challenge in human genomics is to decipher the context specific relationship of sequence to function. However, existing tools for locus specific hypermutation and evolution in the native...
5
47
207
Slide-tags unifies the single cell + spatial genomics worlds. It solves three key problems with existing spatial methods: 1) Cell segmentation (the biggest problem) 2) robustness and scalability (well-powered case-control studies now possible) 3) Multiomics (ATAC, RNA, etc)
A new method called Slide-tags lets scientists capture both genetic and location information of individual cells using standard single-cell workflows in the lab. The technology builds upon Slide-seq, both developed by the labs of @insitubiology & @macosko. https://t.co/2LsxjxZa9L
3
42
275
Discover more in my original preprint thread here: https://t.co/0N7WYURhH2
Single-cell or spatial? Our new technology - Slide-tags - allows both in the same experiment, enabling true single-cell multi-modal spatial genomics ➡️ https://t.co/i1m5T4bEme
0
0
1
This wouldn’t have been possible without a fantastic team: @jacksonweir4*, @Naeemnadaff*, Matthew S, Vipin K, @SandeepKambham2, @RuthRaichur, Gio M, @soph_liu, @KarolBalderrama, Charles V, @Vignesh_Shan, @Luyi_T, Catherine W, Bryan I, Charles Y, @macosko†, @insitubiology†
1
0
9
This was a really fun collaborative project with with: @jacksonweir4*, @Naeemnadaff*, Matthew S, Vipin K, @SandeepKambham2, @RuthRaichur, Gio M, @soph_liu, @KarolBalderrama, Charles V, @Vignesh_Shan, @Luyi_T, Catherine W, Charles Y, @macosko†, @insitubiology†
3
0
9
Please check out the preprint which contains a lot more information, and links to the data! Thank you to everyone at @broadinstitute and @HSCRB who made this possible! @EMBO for funding me.
1
0
7
Slide-tags can profile various macromolecules, tissues, and length scales. Data can be analyzed with existing single-cell workflows too. We believe it will serve as an invaluable tool to study tissue biology.
1
2
4
Additionally, we enriched TCR sequences, and inferred copy number, allowing spatial investigation of the: genetic, chromatin accessibility, transcriptional, and immune, composition of this tumour.
1
0
17
Finally, Slide-tags barcoded nuclei can be profiled with virtually any sn-seq technology. We demonstrated this by performing Slide-tags multimodal snATAC-seq & snRNA-seq on a human metastatic melanoma.
1
0
7
We also profiled: mouse brain at E14, human prefrontal cortex, human tonsil, and human melanoma using Slide-tags snRNA-seq. Slide-tags allowed assignment of gene expression gradients to specific cell types, contextualization of predicted receptor-ligand interactions, and more!
1
1
10
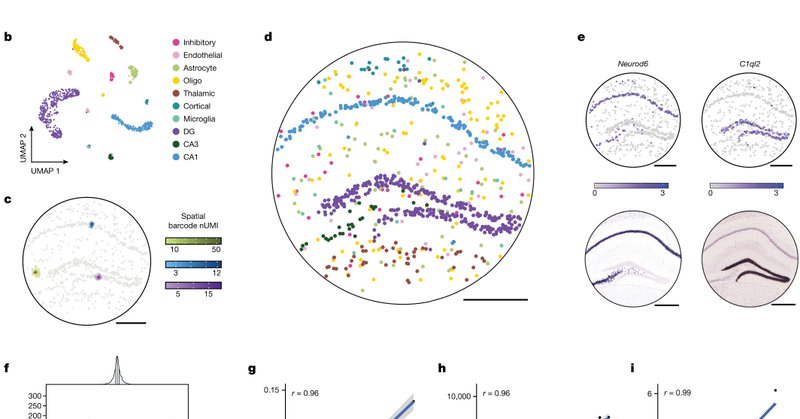
We performed Slide-tags snRNA-seq on the mouse hippocampus to benchmark our technology. Slide-tags recapitulates known tissue architecture, gene expression patterns, and we confirmed that the tagging procedure did not affect the data quality.
1
3
12
In our approach, we “tag” nuclei within intact tissue sections with spatial barcode oligos (whose position is known). After isolation from the tissue, these nuclei can be profiled with virtually any single-nucleus sequencing technology.
1
2
17
Single-cell or spatial? Our new technology - Slide-tags - allows both in the same experiment, enabling true single-cell multi-modal spatial genomics ➡️ https://t.co/i1m5T4bEme
12
168
581
Proud to announce that our paper is out now➡️ https://t.co/HYYZkQ2FV3 We explored how malaria parasites differentiate from asexual replication into sexual cell types ♂️♀️ - please see Theo's excellent 🧵 below for more info and acknowledgements!
Now out in journal form: https://t.co/xWl1SG6LbW Malaria parasites replicate asexually in the bloodstream, however a subset form ♂️ and ♀️ sexual cell types that when sucked up by a 🦟 undergo sexual reproduction. But how? 🧵
1
0
14
Proud to announce that our paper is out now➡️ https://t.co/HYYZkQ2FV3 We explored how malaria parasites differentiate from asexual replication into sexual cell types ♂️♀️ - please see Oliver's excellent 🧵 below for more info and acknowledgements!
0
3
12