Alma Andersson
@aalmaander
Followers
565
Following
660
Media
6
Statuses
147
Senior AI Scientist @ Genentech with a passion for running and the outdoors. All views are my own. She/her 🌈
San Francisco
Joined February 2021
🚨 Alert 🚨 Any1 with an interest in the intersectional space of AI/ML and omics-related stuff should have a look at this workshop 👉 It's designed to bring people from both domains together and has a stellar committee! I'm super stoked 🙌🙌.
mlgenx.github.io
MLGenX plans to bridge the gap between AI and (functional) genomics, with a primary emphasis on target identification.
1
7
25
RT @molly0xFFF: From yesterday's exhibits in US v. Sam Bankman-Fried:. The prosecution shows that the "insurance fund" that FTX bragged abo….
0
828
0
RT @hoheyn: JOIN OUR TEAM @cnag_eu! . WANTED: Spatial Genomics Team Leader. Help us bringing #singlecell research to the next level! . Pr….
0
21
0
RT @SettyM: Super excited to present our algorithm, Mellon for computing densities in single-cell landscapes.
biorxiv.org
Cell-state density characterizes the distribution of cells along phenotypic landscapes and is crucial for unraveling the mechanisms that drive cellular differentiation, regeneration, and disease....
0
36
0
RT @bayraktar_lab: Pls RT! We are hiring a senior computational biologist to work on large sc/spatial datasets of human brain cancer. High….
0
87
0
Not sure what to do this summer? Join our AI/ML team here at @genentech in beautiful SF as an intern! If you're interested in method dev. for spatial-omics or phenotypic screening, we have two kick-ass projects where some extra hands are needed! 🤩.
0
8
40
RT @camillaengblom: Are you, like us, wondering WHERE’S that CLONE? On behalf of the ‘Spatial VDJ’ team, here's full-length BCR and TCR map….
0
64
0
This is a must read for anyone interested in spatial omics! 🤩 To me, this is a landmark paper and an example of really intelligent use of spatial data. Beautiful work, I've been waiting on this since the preprint - congrats to everyone involved!🥳.
Excited that our work on learning cell-cell communication in spatial transcriptomics is finally out @NatureBiotech! Led by @davidsebfischer & @AnnaCSchaar, we setup a graph neural network to estimate niche composition on expression in unbiased fashion.
2
2
68
RT @g_palla1: We've also just published new squidpy release, with several bugfixes for readers and new tutorials for @nanostringtech cosmx,….
0
7
0
RT @PyTorch: Today we’d like to highlight features from functorch, a beta PyTorch library that provides JAX-inspired function transformatio….
0
129
0
RT @scverse_team: We are very excited to announce scverse (, a new consortium around the core Python packages for s….
scverse.org
Foundational tools for single-cell omics data analysis
0
225
0
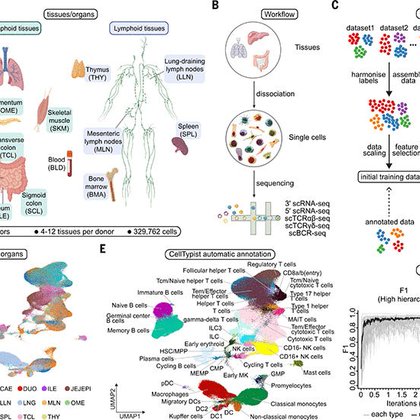
RT @CDominguezConde: 🎉🎉 Extremely proud and grateful that our Tissue Immune Cell Atlas is out in @ScienceMagazine .
science.org
An immune cell atlas of human innate and adaptive immune cells across lymphoid, mucosal, and exocrine sites reveals tissue-specific compositions and features.
0
128
0
A question for all the #spatialomics people, what's your favorite term for the spatial locations and why? I see so many different options: bead, spot, feature, pixel, gexel, etc. Can't we just agree on one?😤🙏. Some thoughts👉
8
3
29
RT @g_palla1: squidpy 1.2 is now available for download! This release features static plotting for diverse spatial technologies, reading fu….
pypi.org
Spatial Single Cell Analysis in Python
0
23
0
RT @singlecellomics: 📢Workshop: "Spatial transcriptomics data analysis in Python" . 💬Speakers tagged on picture. 🔧Tools: squidpy, ncem, cel….
0
50
0
RT @EChelebian: Ever wanted to visualize the results of your experiments on top of full resolution images directly in #JupyterNotebook?. No….
0
3
0
RT @bayraktar_lab: JOB alert. Help us find a staff scientist to map cell states in glioblastoma with sc/spatial omics. Fantastic post for a….
0
99
0
RT @MDLuecken: If you need access to lots of single-cell data for benchmarking & prototyping methods, check out this workshop. It's also an….
0
19
0