Zach Wu
@ZvxyWu
Followers
1K
Following
3K
Media
3
Statuses
383
RS at Google Deepmind, previously Arnold/DeLisa research groups at Caltech/Cornell.
Mountain View, CA
Joined March 2018
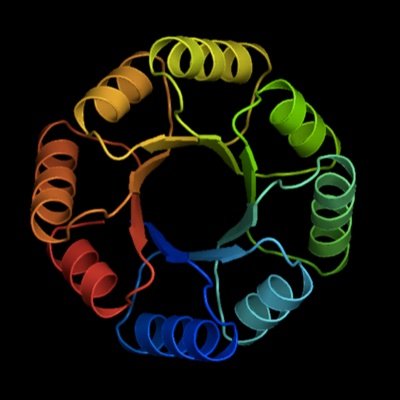
Directed Evolution (DE) is an optimization algorithm for proteins, so Machine Learning (ML) should help! We show on a large fitness landscape that DE+ML improves proteins better than DE alone and provide an example application. https://t.co/QwUPD5ceFZ
@francesarnold @sbjennykan
arxiv.org
To reduce experimental effort associated with directed protein evolution and to explore the sequence space encoded by mutating multiple positions simultaneously, we incorporate machine learning in...
3
48
159
LLM engineered carbon capture enzymes have officially been produced. The best designs were 170% more active and 25% more stable across extreme pH (Tm +8.5 C). Winning strategies include adapting a tag from a bacterial carbonic anhydrase, beta barrel core packing, and removing
31
80
431
Excited to share Caliby 🐈, our new model for structure-conditioned sequence design! Caliby is a Potts model-based sequence design method that can condition on structural ensembles. We use this to average out non-structural signal (e.g. evolutionary bias) learned by models 🧵1/N
3
49
239
Very pleased to see our AI model GenSLM designing novel and versatile enzymes in a challenging setting in @francesarnold lab. https://t.co/0zjpIg716I Back in 2022, we built the world's first genome-scale language model (GenSLM) trained on all known bacterial and viral genomes
3
30
119
Check out our new perspective "Illuminating the universe of enzyme catalysis in the era of artificial intelligence" now out in @CellSystemsCP ! We discuss a vision and path forward for genetically encoding almost all chemistry, powered by new AI tools: https://t.co/Tc7zQ6TExm
3
29
111
Excited to share our recent work: Sparse autoencoders uncover biologically interpretable features in protein language model representations now in PNAS. Thread below 🧵
4
25
108
We have a new collection of protein structure generative models which we call Protpardelle-1c. It builds on the original Protpardelle and is tailored for conditional generation: motif scaffolding and binder generation.
2
29
163
DynaRepo: The repository of macromolecular conformational dynamics 1. DynaRepo is a novel repository that addresses the critical gap in studying the dynamic behavior of macromolecules, which is essential for understanding interactions like antibody–antigen recognition and
0
31
109
🚨New paper 🚨 Can protein language models help us fight viral outbreaks? Not yet. Here’s why 🧵👇 1/12
1
37
155
We made it on the cover! So proud of the team 🎉 You can use BioEmu for free now to get AI predictions for the entire distribution of conformations a protein can take:
github.com
Inference code for scalable emulation of protein equilibrium ensembles with generative deep learning - microsoft/bioemu
Researchers have developed a #DeepLearning system called BioEmu that rapidly generates diverse protein conformations, enabling fast, accurate insights into protein flexibility and function. Learn more this week in Science: https://t.co/Pe15hm9F52
7
35
242
1/ 🧵Can protein language models reason about evolution, not just model it? We built a 24 M-param PLM, Phyla, that reconstructs phylogenetic trees better than existing PLMs. Details & links in 2/👇
5
55
277
Really enjoying #Repartzymes Repurposed and Artificial Enzymes meeting organized by #TomWard of Uni. Basel. Many former group members are here! https://t.co/P3GoQqgUce
1
11
151
Excited to share #AlphaGenome, a start of our AlphaGenome named journey to decipher the regulatory genome! The model matches or exceeds top-performing external models on 24 out of 26 variant evaluations, across a wide range of biological modalities.1/6
13
210
906
🎉New preprint!🎉 Extremely excited to share CryoBoltz❄️⚡️, led by superstar @rishwanth_raghu! We develop a multiscale guidance recipe to steer structure prediction models (e.g. AlphaFold3 / Boltz-1) towards experimental cryo-EM density maps, including heterogeneous,
Excited to present CryoBoltz ❄️⚡, a multiscale guidance approach for steering AlphaFold3/Boltz-1 to sample structures that are consistent with experimental cryo-EM density maps. 🧵1/7 https://t.co/rPMtaPMsGF Joint work with @axlevy0 @GordonWetzstein & @ZhongingAlong!
1
30
209
Excited to unveil Boltz-2, our new model capable not only of predicting structures but also binding affinities! Boltz-2 is the first AI model to approach the performance of FEP simulations while being more than 1000x faster! All open-sourced under MIT license! A thread… 🤗🚀
49
415
2K
🚀 Excited to release BoltzDesign1! ✨ Now with LogMD-based trajectory visualization. 🔗 Demo: https://t.co/tjBXSibZhT Feedback & collabs welcome! 🙌 🔗: GitHub: https://t.co/HsQBrB8amQ 🔗: Colab: https://t.co/TzX4q5m2qp
@sokrypton @MartinPacesa
7
71
365
Guide your favorite protein generative model with experimental data? Meet ProteinGuide - a method to condition pre-trained models on properties without retraining. We validated it both in silico by guiding ProteinMPNN and ESM3 on 3 tasks and in vitro by engineering base editors.
5
40
219
I'm excited to share our new preprint "Steering Generative Models with Experimental Data for Protein Fitness Optimization" (🧵1/5)! Paper: https://t.co/SeOO3QNimY Code:
github.com
Steering discrete diffusion models with labeled data for protein fitness optimization - jsunn-y/SGPO
6
27
138
In a medical milestone, a customized base editor was developed, characterized in human and mouse cells, tested in mice, studied for safety in non-human primates, cleared by @US_FDA for clinical trial use, manufactured as a complex with an LNP, and dosed into a baby with a severe,
44
434
2K
Check out this work led by the incredible @AdaFang_ ATOMICA constructs representations of molecular interactions at the atomic scale, capturing universal physiochemical principles. Read about the exciting insights from applying ATOMICA below 👇
Introducing ATOMICA 💫 A model to universally represent molecular interactions (for proteins, nucleic acids, small molecules, and ions) at an all-atom scale 🧵
0
2
5
Happy Sunday to all! This morning, we are excited to share Chase’s work developing a simple, scalable method to assemble 100s-1000s of custom genes from oligo pools using standard lab tools! (Small 🧵 below)
5
51
189