Cell Systems
@CellSystemsCP
Followers
7K
Following
1K
Media
281
Statuses
3K
A monthly, peer-reviewed journal from Cell Press devoted to Systems Biology. Posts from the Editors. We are on Bluesky @cp-cellsystems.bsky.social
Cambridge, MA
Joined October 2014
Together with our partners @ZJU_China, Liangzhu Laboratory, we're thrilled to announce @CellSymposia Taking programmed cell death from mechanisms to interventions 📍Hangzhou, China 📆October 22–24, 2026 Visit our website: https://t.co/XZNqRzh59V
#CSCellDeath26
0
0
0
Hear @EimearEKenny share her work “From categories to context: Rethinking ancestry for precision medicine” @CellSymposia #CSPrecisionGen25, Sept 17–19, 2025 San Diego, CA. 🔗Register here: https://t.co/NrS8jysr50
1
0
0
The picks and shovels of the AI economy. AIS provides exposure to companies at the forefront of artificial intelligence—spanning semiconductors, data centers, and AI-enabled applications. Consider how AIS may align with your investment strategy.
0
12
94
Excited to hear @uarizona’s @DavidEnard share insights on “The genetic legacy of ancient viral epidemics in human host genomes” @CellSymposia #CSPrecisionGen25. Don’t miss it—register today! 🔗 https://t.co/8XM3lKLd0m
1
0
0
Biotechnology plays a crucial role in areas such as healthcare, agriculture, environmental sustainability, and industrial processes. Journals across Cell Press have come together to share exciting papers in this growing field. Visit the collection: https://t.co/t5CyZGs9QD
0
0
0
We were so excited to see the innovative science at @KeystoneSymp's Machine Learning Applied to Macromolecular Structure and Function! Access the content now on-demand: https://t.co/cfYlVXIrWm
#KSMLStructure25
0
1
2
1/ „What is the main bottleneck in deriving biological understanding from spatial transcriptomic profiling?" @CellSystemsCP asked a few of us for comments on this, I think quite cool read here at
4
10
71
We asked @MoffitLab, @DrMingyaoLi, Qing Nie @uci_cmcf, @kazukane, @teichlab, Daniel Dar @WeizmannScience, @ltkagohara, @SItzkovitz, @roserventotormo, @ItaiYanai, @FertigLab, and @fabian_theis about the main bottlenecks facing Spatial Transcriptomics
cell.com
Image-based approaches to single-cell transcriptomics offer the exciting ability to directly image gene expression at the transcriptome scale within intact tissue slices, defining, discovering, and...
0
10
38
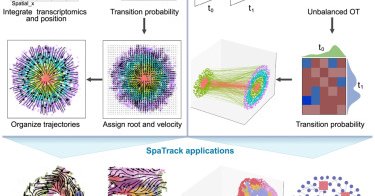
Shen et al. introduce SpaTrack to reconstruct cell differentiation trajectories from spatial transcriptomics data at single cell resolution within a spatial context by using optimal transport #BGI-Research @BGI_Genomics
cell.com
Shen et al. introduce SpaTrack, which integrates cell transcriptomics and spatial position using optimal transport to reconstruct cell differentiation trajectories. SpaTrack is applied to disentangle...
0
0
2
Our February issue is online. On the cover: Regenerative trajectories of axolotl telencephalon regeneration inferred by SpaTrack using spatial transcriptomic data. https://t.co/3sbMgnaLoP
1
0
7
🚨 Early Career Researchers 🚨 Don’t miss out on the Alma Dal Co School on Collective Behaviour (Sep 29 - Oct 4, 2025, Venice, Italy)! 🧬🌍 An interdisciplinary event with talks from 18 world-class experts #qbio! 🤝💡 ⏳ Deadline: Feb 28, 2025 Apply now:
almadalcoschool.github.io
The Alma Dal Co School on Collective Behaviour
0
3
9
"Scientists need to re-imagine their role in society, incorporating new models of public service & mutual learning with communities to ensure science remains relevant, impactful & transformative for society." - @Thumbi_Ndungu for @ImmunityCP 'service' ➡️ https://t.co/WTkKRQXpgy
1
7
16
Get to know TQQQ: The world’s largest leveraged ETF, targeting 3x daily Nasdaq-100 returns.*
0
20
200
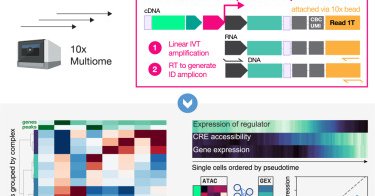
Metzner et al. present Multiome Perturb-seq, extending single-cell CRISPR screens to simultaneously quantify perturbation effects on the epigenome and transcriptome in a scalable way, describing the integrated effects on cell state @thenormanlab
cell.com
Multiome Perturb-seq extends single-cell CRISPR screens to simultaneously quantify perturbations’ effects on the epigenome and transcriptome. A CRISPRi screen of chromatin remodelers using this...
0
1
1
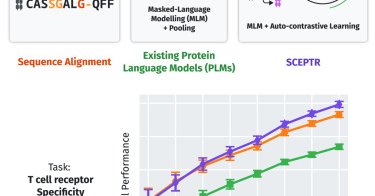
Nagano et al. present SCEPTR , a TCR language model with a pre-training strategy combining autocontrastive learning and masked-language modeling, to predict TCR specificity, outperforming existing language models and sequence-based methods @andimscience
cell.com
How should we train protein language models to predict protein function? This study shows that pre-training using an autocontrastive loss greatly improves transfer learning compared with masked-lan...
1
7
37
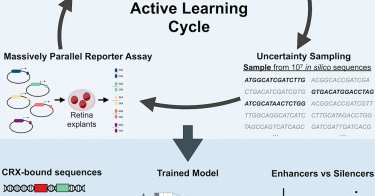
Using active learning with MPRAs in the developing mouse retina, @rfriedman22 et al. train a CNN to distinguish between activating and repressing regulatory DNA elements composed of the same TF binding sites @genologos @CorboLab @WashUGenetics
cell.com
Friedman et al. introduce an active machine learning workflow to iteratively train deep learning models of regulatory DNA on successive rounds of experimental data. Using active learning with...
0
1
5
From the builders coding the next protocol to the institutions allocating capital, everyone who matters in Web3 gathers at Consensus. Join thousands of attendees in Miami for 3 days of collaboration, learning, and partnership building. It’s the one event you can't afford to miss.
0
0
0
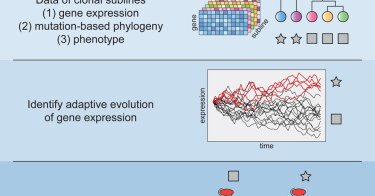
Hirsch et al. use a stochastic model of gene expression evolution to study adaptation in melanoma subclones and uncover gene expression patterns associated with canonical and non-canonical Wnt signaling resulting in the proliferative & invasive phenotypes
cell.com
This study uses a stochastic model of gene expression evolution to study evolutionary adaptation in melanoma subclones with distinct phenotypes. It uncovers distinct adaptive gene expression patterns...
1
1
2
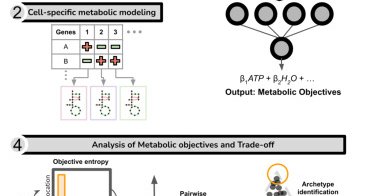
Lin et al. use machine learning and metabolic modeling to infer cellular objectives from multi-omics data, revealing trade-offs between stress protection and growth in embryonic development @sriram_lab
cell.com
How do cells balance competing metabolic needs? This study introduces SCOOTI, a tool to infer objectives and trade-offs in cellular metabolism using machine learning and modeling. SCOOTI revealed...
0
1
2
Hoffman et al. demonstrate that a common ERK kinase translocation reporter (KTR) is also activated by CDK2, develop computational correction methods to subtract CDK2 signal, and present a new ERK KTR that does not show crosstalk with CDK2. @Spencer__Lab
0
0
0
Our first issue of 2025 is online! On the cover: Heatmap of temporal activity of ERK, measured using the modified ERK kinase translocation reporter and read out as cytoplasmic:nuclear ratio in single cells before and after treatment with 10 nM Trametinib. https://t.co/CUmFA93oyp
2
1
4
Join our editor Dr. Ernesto Andrianantoandro at @KeystoneSymp's, Machine Learning Applied to Macromolecular Structure and Function. Scholarship, abstract submissions, and early registration are still open! Register today: https://t.co/sFyp7yufvh
#KSMLStructure25
0
1
4