Honggui Wu
@Wu_Honggui
Followers
825
Following
7K
Media
24
Statuses
490
Postdoc in Feng Zhang lab @BroadInstitute. Graduated from Sunney Xie Lab at Peking University.
Boston
Joined September 2017
Whole-genome single-cell multimodal history tracing to reveal cell identity transition https://t.co/evdWPaIsS4
#biorxiv_genomic
biorxiv.org
Advances in single-cell sequencing have deepened our understanding of cellular identities. However, because they inherently capture only static snapshots, after which no further observations are...
0
6
20
Spatial methylome + transcriptome.
Spatial joint profiling of DNA methylome and transcriptome in mammalian tissues https://t.co/TvIPN4DRFM
#biorxiv_genomic
0
1
6
Our scMicro-C paper is out in @NatureGenet! Huge thanks to the reviewers for their valuable insights, to @XieSunney for unwavering support, and to fantastic collaborators @JiankunZhang21 & @tanlongzhi. Thrilled to share this work!
Very excited to share our work on high-resolution single-cell Micro-C, which is on bioRxiv now. Here we reveal that cohesin-mediated loop extrusion participates in transcription with kilobase-resolution single-cell 3D structures.
3
17
94
Ironman. Maternal iron deficiency causes male-to-female sex reversal in mouse embryos https://t.co/dgnjuHPfV3
@Nature
4
42
206
In @nature we describe the use of scalable #proteinengineering & #machinelearning to predict millions of bespoke CRISPR-Cas9 enzymes, offering safer & more genome editing tools. 🧬🖥️ @RachelSilvers11 @CGM_MGH @MGH_RI @MassGeneralNews @harvardmed
https://t.co/oNtG93uXCl
nature.com
Nature - Combined high-throughput protein engineering with machine learning to curate libraries of CRISPR genome-editing enzymes with distinct genome targeting properties is described.
41
72
262
🔥🔥🔥OUR NEW PREPRINT: Slowing and reversing aging? Nature does it, here we show how! Age deceleration and reversal gene patterns in dauer diapause. Huge congrats to Kristy Totska, João Barata, Walter Sandt and @Meyer_DH
biorxiv.org
The aging process is characterized by a general decrease in physical functionality and poses the biggest risk factor for a variety of diseases such as cancer, cardiovascular diseases, and neurodege...
10
33
144
Introducing RAEFISH, our lab's new flagship image-based spatial transcriptomics technology that simultaneously enables single-molecule spatial resolution and whole-genome level coverage of long and short, endogenous and engineered RNA species in cell cultures and intact tissues.
1
55
276
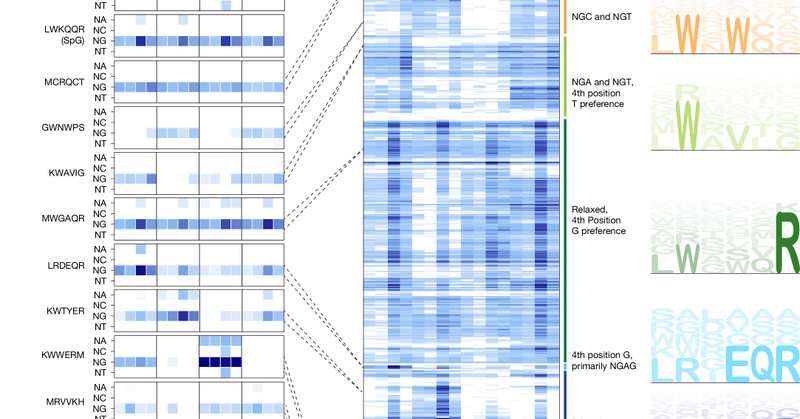
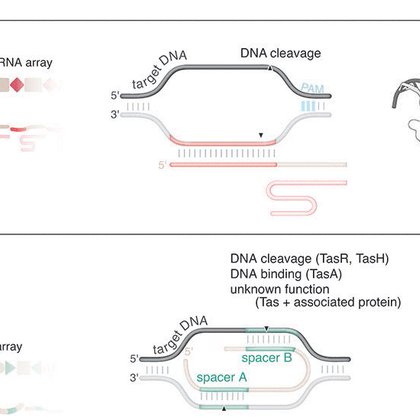
We are excited to report the discovery of TIGRs, a widely-occurring RNA-guided system found in bacteria and their viruses. TIGRs consist of a peculiar repeat region which is transcribed into RNA and processed into multiple guide RNAs to direct TIGR-associated proteins to their
science.org
RNA-guided systems provide remarkable versatility, enabling diverse biological functions. Through iterative structural and sequence homology-based mining starting with a guide RNA-interaction domain...
26
271
1K
Sequencing by Expansion (SBX) -- a novel, high-throughput single-molecule sequencing technology https://t.co/kpve9uP9vp
#biorxiv_genomic
biorxiv.org
Remarkable advances in high-throughput sequencing have enabled major biological discoveries and clinical applications, but achieving wider distribution and use depends critically on further improve...
1
17
66
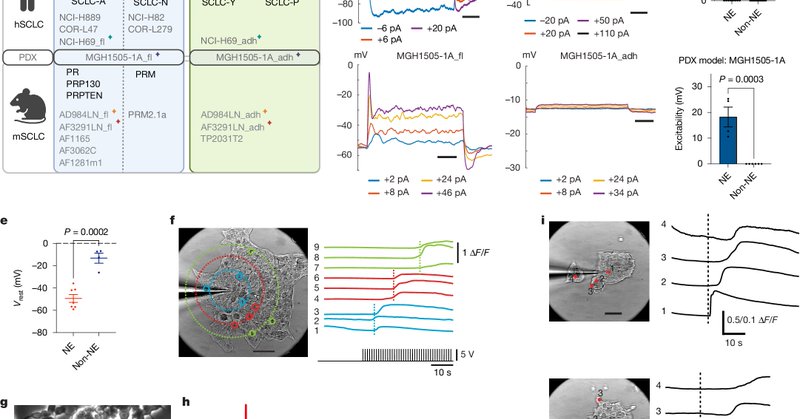
Did you know cancer cells can be electrically active, just like neurons? This electrical activity plays a key role in tumor progression. Excited to share my first postdoc paper at @TheCrick (and first paper from @LiGroup_Crick), published in @Nature! https://t.co/bqekzffnG0
nature.com
Nature - Electrical excitability in neuroendocrine SCLC cells promotes tumour progression through action potential firing, increasing ATP demand and oxidative phosphorylation dependency, whereas...
10
69
283
New #preprint from our lab at @IMBA_Vienna ! How do DNA breaks locate homology sites in the vast space of the human genome? We show how #cohesin guides homology search for faithful repair! Read more 👉 https://t.co/ouPmrddhPr Follow along for key insights! 🧵1/12
1
13
35
.@LanJiangBoston and colleagues developed droplet-based UDA-seq by incorporating a post-indexing step into two-round combinatorial indexing, enhancing throughput for large-scale single-cell analysis. https://t.co/j4XFlSKtXA
0
5
29
It has been a wonderful collaboration with @MaoxuC, and I deeply appreciate @XieSunney 's support and guidance throughout the project.
0
0
3
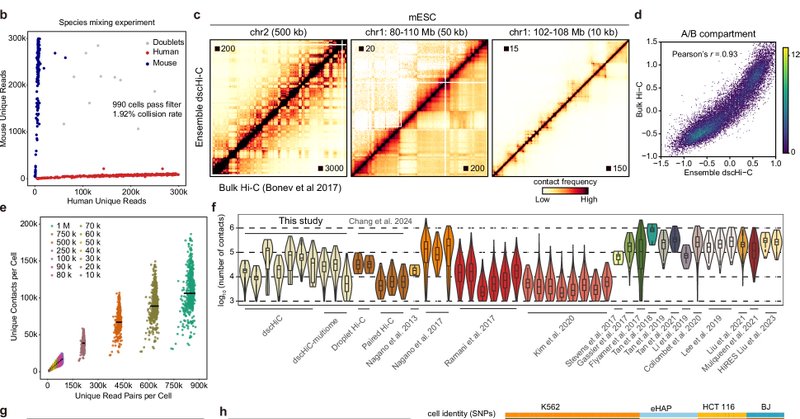
dscHi-C allows us to investigate chromatin reorganization during mouse brain aging, while dscHi-C-Multiome reveals the complex relationship between genome structure and gene regulation. Learn more here: https://t.co/NdDtS7Byqg
nature.com
Cell Discovery - Droplet-based high-throughput 3D genome structure mapping of single cells with simultaneous transcriptomics
1
1
3
Thrilled to introduce a new single-cell 3D genome analysis tool to our toolkit! Our high-throughput droplet Hi-C (dscHi-C) and dscHi-C-multiome methods use droplet microfluidics to generate tens of thousands of single-cell Hi-C profiles with high sensitivity in a single batch.
5
27
150
WHY DOES THE EPIGENETIC CLOCK TICK? While money has been poured into developing therapies to reverse the epigenetic clock (@altos_labs 🤑), why the epigenome changes with age at all is unclear. In @NatureAging today, we suggest a potential driver: somatic mutations.
15
212
995
Congratulations!
After my first public report at the Double Helix 70’s celebration ( https://t.co/XDsJIdxYFE) on Oct 2023 and a bioRxiv on June 2024, FOOtprinting by dsDNA DeamInasE (FOODIE), the major effort of my CPL/PKU group, delayed by the pandemic, has been yearmarked by today issue of PNAS
0
0
10
A foundation model for enzyme retrieval and function prediction is released. EnzymeCAGE is trained on ~1M enzyme-reaction pairs via CLIP, and can be used to annotate unseen enzymes or orphan reactions. We open-source it before releasing the manuscript:
github.com
Contribute to GENTEL-lab/EnzymeCAGE development by creating an account on GitHub.
2
17
124
Have you ever wondered how your back formed? The human embryo makes a neural tube (future spinal cord) and somites (trunk muscle/bone) from ~d20. They’re formed at the same time and place, so we used human Trunk-like Structures (hTLS) to investigate their ‘co-development’… (1/7)
10
43
233
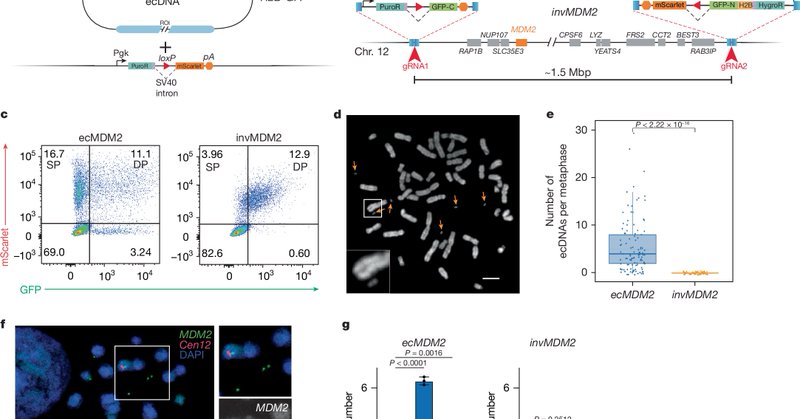
1/ 🚨New paper on ecDNAs from our lab! We reveal a strategy to engineer extrachromosomal DNA (ecDNA) amplifications in cells & mice. Let's dive in! 🧵👇 https://t.co/ct2s4Rhcty
nature.com
Nature - Large extrachromosomal DNAs are engineered using a CRISPR- and Cre–loxP-based approach and shown to drive cancer in mouse models, with potential applications in determining the role...
27
91
318