Tony
@Tony_Chen6
Followers
103
Following
67
Media
14
Statuses
32
Postdoc @MGHMedicine / @BroadInstitute || PhD from @HarvardBiostats
Joined March 2020
We’re excited to share our latest publication in @CellGenomics: “Streamlining Large-Scale Genomic Data Management: Insights from the UK Biobank Whole-Genome Sequencing Data”. Sincerely thanks to @drarwood, @muzizimumu1, @XihongLin, Yuxin Yuan, Gareth Hawkes, Robin Beaumont,
Streamlining large-scale genomic data management: Insights from the UK Biobank whole-genome sequencing data
1
13
57
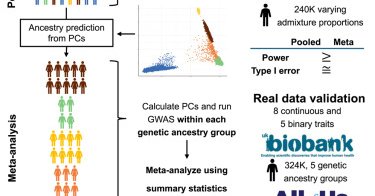
So excited to share my first PhD paper, discussing GWAS methodologies in multi-ancestry contexts: To pool or not to pool? Very proud of this work and grateful for the supervision of @AndrewHaoyu and @GENES_PK! Read more:
cell.com
Multi-ancestry GWASs enhance discovery in diverse populations, but optimal methods remain debated. Using theory, simulations, and analyses from the UK Biobank and All of Us, we show that pooled...
2
5
19
Excited to share our new manuscript on evaluating multi-ancestry GWAS methods using large-scale simulations & real data from UK Biobank (N≈324K) & All of Us (N≈207K)! My first thesis paper under the guidance of @GENES_PK & @AndrewHaoyu. Available at: https://t.co/xxAbJXn7zZ
1
6
30
Please join us tomorrow at HSPH for a @HarvardPqg Working Group seminar with @kunhsingyu , who will be discussing AI foundation models in pathology!
0
0
6
📢Out now! @xihaoli, @pnatarajanmd, @muzizimumu1, @ZHLiu13, @XihongLin, and colleagues from @nih_nhlbi present a framework for functionally-informed multi-trait rare variant analysis of biobank-scale sequencing studies. https://t.co/judklgK1eO 🔓 https://t.co/0ua7YW6Dko
2
10
34
Please join us for our last @HarvardPqg Working Group seminar of 2024, next Tuesday with @yk_tani presenting his exciting recent work on polygenic prediction!
I'm excited to present at the Program in Quantitative Genomics @HarvardPqg Working Group seminar next Tuesday. It's always a pleasure to receive seminar invitations from students. Thank you, @Tony_Chen6. I'm looking forward to it!
0
1
7
Just in time! Come join us with @saorisakaue on 11/19 for her recent work revealing genetic regulatory mechanisms through eQTL causal variants! @HarvardBiostats @HarvardEpi
0
2
13
Our paper introducing a PRS-based translation roadmap for lung cancer care is now published on @eBioMedicine ! Wonderful collaboration with @AndrewHaoyu , @lishiunchen , ILCCO and GSCAN towards the clinical implementation of PRS. https://t.co/pLqPpEzj6k
Very excited to share our preprint presenting a framework for translation of PRS to primary care for lung cancer in diverse high-risk patients. Grateful for the support from @AndrewHaoyu and @lishiunchen, and amazing collaborators from WashU and ILCCO. https://t.co/MzLHuh32Mi
0
1
8
🚀 Excited to share our new preprint on RICE, a PRS framework that integrates common and rare variants to enhance genetic risk prediction across diverse ancestries! Led by my postdoc fellow Jacob Williams, co-mentored with @xihaoli. Check it out here: https://t.co/0VbsFchE1Q 1/N
1
15
78
Check out the presentations from our lab and our close collaborators #ASHG2024. Such a hard-working year. So proud of the group!!! Very humbled to be able to work with so many great collaborators @xihaoli @XihongLin @GENES_PK @Diptavo @HongyuZhao2 @Tony_Chen6 @AlexiaDiasF
1
5
23
Just in time, I'll be presenting this work today at the @HarvardPqg conference in Boston, and at the @GeneticsSociety conference in Denver. Looking forward to comments, feedback, and discussion!
SPLENDID enables computation of a single, label-free model with robust prediction across all ancestries, which we hope can enhance fairness and interpretability in clinical implementation. Software and tutorials are available on GitHub. https://t.co/FnmT06667n
0
0
9
SPLENDID enables computation of a single, label-free model with robust prediction across all ancestries, which we hope can enhance fairness and interpretability in clinical implementation. Software and tutorials are available on GitHub. https://t.co/FnmT06667n
Using training data from All of Us and validation in UK Biobank, SPLENDID outperforms existing methods, particularly in non-European ancestry, selects fewer variants, remains robust to ancestry proportions in tuning, and can be used with only training data.
0
0
3
Using training data from All of Us and validation in UK Biobank, SPLENDID outperforms existing methods, particularly in non-European ancestry, selects fewer variants, remains robust to ancestry proportions in tuning, and can be used with only training data.
SPLENDID uses an ensemble group-L0L2 penalized regression framework to model interactions between genetic variants and ancestry PCs. This allows for simultaneous estimation of shared and heterogeneous effects across diverse populations without discrete ancestry labels.
0
0
3
SPLENDID uses an ensemble group-L0L2 penalized regression framework to model interactions between genetic variants and ancestry PCs. This allows for simultaneous estimation of shared and heterogeneous effects across diverse populations without discrete ancestry labels.
Excited to share our preprint for SPLENDID, a new PRS method using continuous ancestry interactions and biobank-scale individual level data. Many thanks to @AndrewHaoyu , Rahul Mazumder, and @XihongLin for their valuable guidance on this work! https://t.co/vb2i6lRekA
0
0
2
Excited to share our preprint for SPLENDID, a new PRS method using continuous ancestry interactions and biobank-scale individual level data. Many thanks to @AndrewHaoyu , Rahul Mazumder, and @XihongLin for their valuable guidance on this work! https://t.co/vb2i6lRekA
0
6
24
Excited to have you @Aoxing2 , really looking forward to your talk!!
Hi - I will give a seminar about our recently published Nature paper on “mosaic X chromosome loss” at the PQG working group NEXT TUESDAY! Welcome to Join the talk and discussion in person!! ⏰Time: 1-2 PM, Tuesday, Oct 22 🏠Location: Biostats Conference Room (2-426), Harvard
1
0
5
Our new paper on semi-supervised machine learning method for predicting homogeneous ancestry groups to assess HWE in diverse whole #genome sequencing studies. The work was motivated by NHGRI Genome Sequencing Program - CCDG data. Applied to HWE QC of 60,000+ diverse whole genomes
📢Online now! 📰Semi-supervised machine learning method for predicting homogeneous ancestry groups to assess Hardy-Weinberg equilibrium in diverse whole #genome sequencing studies 🧑🤝🧑@xihonglin @bmneale @mczody & co https://t.co/0rAce5EFkM
1
6
21
ALL-Sum combines fast optimization of L0L2 penalized regression with ensemble learning to improve PRS accuracy and computational efficiency vs. many popular methods. Software and accompany data are all available on GitHub ( https://t.co/ONXJnh1uzN)
0
0
3
Our paper introducing a fast and scalable PRS method (ALL-Sum) is now available on @PNASNews! Many thanks to @AndrewHaoyu, Rahul Mazumder, and @XihongLin for their mentorship, as well as @BhramarBioStat and @HongyuZhao2 for reviewing! https://t.co/jx4fW4Km1P
1
2
27
Genomic Insights for Personalized Care: Motivating At-Risk Individuals Toward Evidence-Based Health Practices https://t.co/riP58pPers
#medRxiv
0
3
1