Hongyu Zhao
@HongyuZhao2
Followers
574
Following
714
Media
3
Statuses
190
Joined February 2018
RT @pnatarajanmd: Delighted to collaborate with J. Hu and @HongyuZhao2 to show that genetic mechanisms leading to a high CAD polygenic risk….
0
9
0
RT @MattGirgenti: Very proud of our new #SingleCell work on #PTSD and #MDD out in @Nature today. Paper: News and v….
github.com
Contribute to mjgirgenti/PTSDsnDLPFC development by creating an account on GitHub.
0
11
0
RT @MattGirgenti: Huge thanks to all coauthors including @HongyuZhao2 @JingZhang_bio @MarkGerstein @kristenbrennand. Special thanks to @Yal….
0
1
0
RT @sgqcheng: scMODAL is a deep learning framework that leverages known feature links to align single-cell omics. @GefeiWang001 @HongyuZhao….
nature.com
Nature Communications - Single-cell multi-omics integration is challenged by varied feature relationships and modality-specific limitations. Here, the authors present scMODAL, a deep learning...
0
6
0
RT @KavliAtYale: In a new @Yale study, researchers led by @MattGirgenti @YalePsych used a comprehensive multi-omic approach to analyze post….
medicine.yale.edu
Yale scientists, led by senior author Matthew Girgenti, PhD, assistant professor of psychiatry, established a brain multi-omic and multi-region analysis
0
3
0
RT @MattGirgenti: Excited to share our latest study of the PTSD prefrontal cortex out @GenomeMedicine. Our systems….
0
1
0
RT @Yale: The wait is over: It’s Match Day at Yale School of Medicine!. After years of hard work, members of the M.D. Class of 2025 have ju….
0
12
0
RT @clintomics: Nice benchmarking study of 10 different foundation models for single cell data analysis. important best practices, limitati….
0
28
0
RT @meSuper8: We have two great interns, Zhiyuan and Wenxin, who co-lead this project. Thanks to Yuge, Dr. Zhang @lesleylezhang and Dr. St….
0
1
0
Happy to share HBI, a Bayesian method to infer cell-type-specific meQTLs led by Youshu Cheng, together with Dr. Ke Xu and others @ysphbiostat @YaleSPH @YaleMed @YalePsych
link.springer.com
Methylation quantitative trait loci (meQTLs) quantify the effects of genetic variants on DNA methylation levels. However, most published studies utilize bulk methylation datasets composed of differ...
0
4
15
RT @sgqcheng: Authors @leihouyeung @LinYingxin @xin4gao @HongyuZhao2 present SANTO, a coarse-to-fine method for spatial omics data integrat….
nature.com
Nature Communications - Spatial omics technologies require alignment and stitching of slices for a 3D molecular profile. Here, the authors present SANTO, a coarse-to-fine method that rapidly...
0
3
0
Another paper led by Jiangnan on joint modeling of human cortical structure through genetic correlation and composite-trait correlation analysis of UK Biobank data, in collaboration with @YiliangElon, Zhaohan, Youshu, Biao, and Yize. @ysphbiostat @YaleSPH
0
1
11
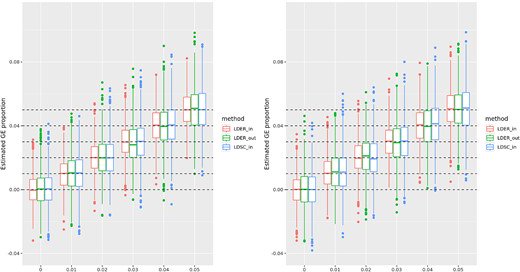
Happy to share the paper led by Zihan Dong on estimating phenotype variance contributed by gene-environmental interactions using GWAS summary statistics, in collaboration with @jiangw90, Hongyu (Eddie) Li, and Andy DeWan @ysphbiostat @YaleSPH
academic.oup.com
Abstract. Gene–environment (GE) interactions are essential in understanding human complex traits. Identifying these interactions is necessary for decipheri
0
3
19
RT @amyytzhao: Our paper on immune profiling pts with Fibrotic Hypersensitivity Pneumonitis is out! . Cannot thank all of our brilliant co….
0
11
0
RT @RichaSharmaMD: Thrilled to share our article about our machine learning algorithm that classifies ischemic stroke etiology using electr….
0
3
0
RT @KaminskiMed: Wow! Yale Basketball beats Auburn!! .Congratulations.@YaleCoachJones .@Daniel_Wolf6 .@BezMbeng @MattKnowling @augustmahon….
0
3
0
Happy to share a review led by @YuhanJustinaXie on assessing de novo variant effects on birth defects with co-authors Ruoxuan Wu, Hongyu Li, Weilai Dong, and @geyu_zhou. @ysphbiostat, @YaleSPH, @YaleMed
link.springer.com
With the development of next-generation sequencing technology, de novo variants (DNVs) with deleterious effects can be identified and investigated for their effects on birth defects such as congeni...
0
3
24