Kun-Hsing Yu
@kunhsingyu
Followers
554
Following
66
Media
8
Statuses
68
Associate Professor, Department of Biomedical Informatics, Harvard Medical School
Boston, MA
Joined May 2012
So thankful, @EkaterinaPeshev and @cat_caruso, for the brilliant news release: https://t.co/1tQUcjQGzT
@harvardmed
hms.harvard.edu
New tool helps surgeons tell apart aggressive glioblastoma from other cancers in the brain
Delighted to share our new paper! We introduce an uncertainty-aware pathology AI that diagnoses cancers with overlapping profiles and knows when it doesn't know. Full article: https://t.co/pWUnojRFtU
@HarvardDBMI @NIH @NIGMS @CDMRP @AmericanCancer #DigitalPathology #AI
0
0
1
@HarvardDBMI @NIH @CDMRP @AmericanCancer Deeply appreciative of our incredible team (2/2): @KeithLigon5 @OmarArnaout12 @ Thomas Roetzer-Pejrimovsky @ Shih-Chieh Lin @ Natalie NC Shih @ Nipon Chaisuriya @ David J. Cook @ Jung-Hsien Chiang @ Chia-Jen Liu @ Adelheid Woehrer @Jeff_golden1 @MacNasrallah
0
0
0
@HarvardDBMI @NIH @CDMRP @AmericanCancer Deeply appreciative of our incredible team (1/2): @ Junhan Zhao @ Shih-Yen Lin @ Raphaël Attias @lizamathewskim @ Christian Engel @ Guillaume Larghero @ Dmytro Vremenko @ Ting-Wan Kao @ Tsung-Hua Lee @ Yu-Hsuan Wang @ Cheng Che Tsai @ElianaMarostica @ Ying-Chun Lo @NeuropathDM
0
0
0
Delighted to share our new paper! We introduce an uncertainty-aware pathology AI that diagnoses cancers with overlapping profiles and knows when it doesn't know. Full article: https://t.co/pWUnojRFtU
@HarvardDBMI @NIH @NIGMS @CDMRP @AmericanCancer #DigitalPathology #AI
nature.com
Nature Communications - Distinguishing glioblastoma and primary central nervous system lymphoma (PCNSL) remains challenging due to their overlapping pathology features. Here, the authors develop a...
2
1
6
Delighted to publish our review article on the generative era of medical AI in Cell! w/ @EricTopol JohnFahrner and Emma Chen
6
124
537
Fun to share our work on Generalist Medical AI, and our applications in GI explored with Dr Tyler Berzin and Romain Hardy
Excited to be at the ASGE AI Summit, exploring just how far AI has come. @pranavrajpurkar @UmaMahadevanIBD
0
2
15
Ever wish you could hit "undo" on disease? 🩺🔄 https://t.co/VW9BSsvJd7 Most drug discovery asks: what does this perturbation do to cells? But we can also ask the reverse: which perturbations undo a disease signature and move cells back toward health? That's the idea behind
8
84
408
Update from CURE-Bench @NeurIPSConf: 900+ entrants and 1,100+ submissions 👉 New teams welcome. Tracks, rules, and leaderboard: https://t.co/D65SR2dmPs
#NeurIPS2025 #CUREBench 💻 Starter kit https://t.co/NQup9mZpFo now includes code + tutorials for @OpenAI GPT-OSS open-weight
Update from CURE-Bench at @NeurIPSConf: 524 entrants and 298 submissions! Thank you to the CURE-Bench community! Working on AI for drug discovery and reasoning in medicine? Your agent belongs here New teams welcome. Tasks, rules, and leaderboard: https://t.co/q40pyydv9C
1
6
24
A huge shout-out to our amazing team: @ XiyueWang @ JunhanZhao @ElianaMarostica @ ChristopherRJackson @ Sen Yang @lmsholl @NeuropathDM @KeithLigon5 @shuji_ogino @Jeff_golden1 @MacNasrallah
0
0
3
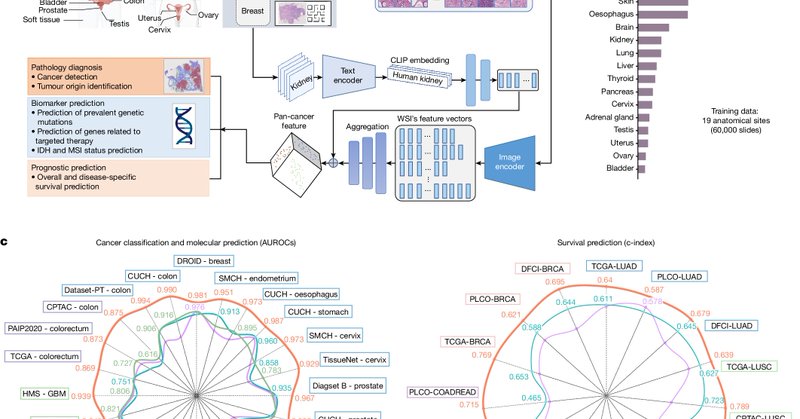
Excited to share that our paper on a foundation AI model for cancer prognosis and pathology diagnosis is published in Nature today. https://t.co/HShXzxVeZE
@HarvardDBMI @NIH @NIGMS @CDMRP @AmericanCancer #AI #DigitalPathology
nature.com
Nature - A study describes the development of a generalizable foundation machine learning framework to extract pathology imaging features for cancer diagnosis and prognosis prediction.
5
26
82
🎉 Excited to share that my NIH R35 grant on causal inference methods for treatment heterogeneity has been funded! Grateful for the support from my mentors and colleagues! 🙌 @NIGMS @PennState @PennStHershey #CausalInference
6
3
40
After benchmarking more than 80 LLMs, @zimmskal & team found that the best model isn't always a great match for your programming language. Google's Gemini Pro 1.5 worked well for Go, but not so much for Java & Ruby, for example. The best overall LLM was Anthropic’s Sonnet 3.5.
4
49
152
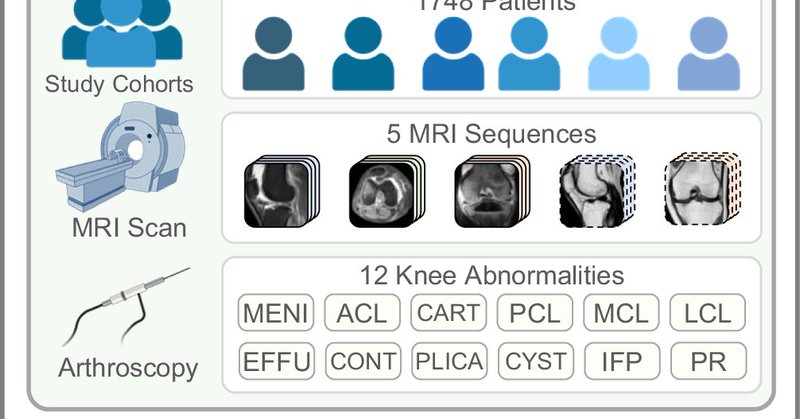
Excited to share our new paper in Nature Communications! Learning co-plane attention across MRI sequences for diagnosing twelve types of knee abnormalities. The diagnosis accuracy of all radiologists was improved significantly with model assistance. https://t.co/dLTMQyYW5c
nature.com
Nature Communications - The authors present a deep learning model that incorporates co-plane attention across image sequences with a performance comparable to senior radiologists in classifying 12...
2
2
18
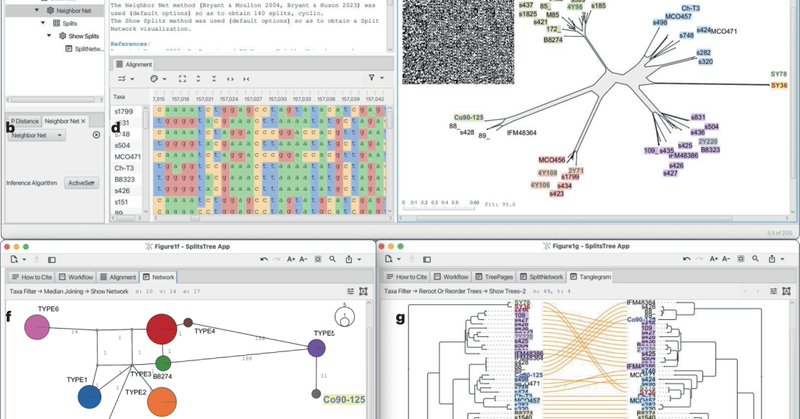
The SplitsTree App: interactive analysis and visualization using phylogenetic trees and networks | Nature Methods
nature.com
Nature Methods - The SplitsTree App: interactive analysis and visualization using phylogenetic trees and networks
0
197
621
We should use soft-max to mean "log of sum of exponentials." What is often called soft-max should really be called soft-argmax. Even John Bridle, who coined the word soft-max, agrees.
The soft-argmax is the gradient of the soft-max (log-sum-exp). Central to perform classification using logistic loss. Needs to be stabilized using the log-sum-exp trick. https://t.co/t2sANAWsLZ
https://t.co/n0Jalhbm3d
33
72
707
Our lab’s new work that shows deeper integration of vision and robotic learning! 🤩🦾
What structural task representation enables multi-stage, in-the-wild, bimanual, reactive manipulation? Introducing ReKep: LVM to label keypoints & VLM to write keypoint-based constraints, solve w/ optimization for diverse tasks, w/o task-specific training or env models. 🧵👇
17
51
343
🚀 Beyond excited to announce our release of the #Cell2Sentence (C2S) API and new foundation models! 🎉 Our C2S API makes it incredibly easy to convert #singlecell data into cell sentences, perform inference with LLM-based C2S models, fine-tune them, and convert cell sentences
7
63
248