Haoyu Zhang
@AndrewHaoyu
Followers
568
Following
329
Media
47
Statuses
376
Stadtman tenure-track investigator at the National Cancer Institute. Statistics, Genetics and Genomics, Data Science. Views are all mine
Cambridge, MA
Joined July 2016
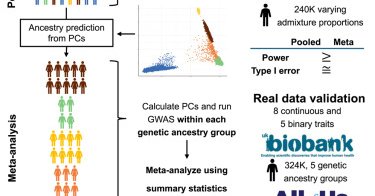
🚨New today! 📰Evaluating multi-ancestry #GWAS methods: Statistical power, population structure, and practical implications 🧑🤝🧑@AlexiaDiasF @AndrewHaoyu @GENES_PK & co https://t.co/EpPDORudan
cell.com
Multi-ancestry GWASs enhance discovery in diverse populations, but optimal methods remain debated. Using theory, simulations, and analyses from the UK Biobank and All of Us, we show that pooled...
0
3
15
This paper is now out today! @AJHGNews
cell.com
Multi-ancestry GWASs enhance discovery in diverse populations, but optimal methods remain debated. Using theory, simulations, and analyses from the UK Biobank and All of Us, we show that pooled...
Excited to share our new manuscript on evaluating multi-ancestry GWAS methods using large-scale simulations & real data from UK Biobank (N≈324K) & All of Us (N≈207K)! My first thesis paper under the guidance of @GENES_PK & @AndrewHaoyu. Available at: https://t.co/xxAbJXn7zZ
2
6
19
Designing a multi-ancestry GWAS? Curious about approaches for association analyses? Check out our new paper in AJHG showing that pooled analyses can offer higher power than meta-analyses. A great collaboration with @AlexiaDiasF @GENES_PK @Tony_Chen6 @madduri!
So excited to share my first PhD paper, discussing GWAS methodologies in multi-ancestry contexts: To pool or not to pool? Very proud of this work and grateful for the supervision of @AndrewHaoyu and @GENES_PK! Read more:
0
3
18
Genetic and Cellular Architecture of Breast Cancer Risk in Multi-Ancestry Studies of 159,297 Cases and 212,102 Controls
medrxiv.org
Breast cancer genome-wide association studies (GWAS) have identified over 200 independent genome-wide significant susceptibility markers. However, most studies have focused on one or two ancestral...
0
1
6
Excited to share our paper, Health risks and genetic architecture of objectively measured multidimensional sleep health, led by Shengkui Zhang, Manrui Zhang, and Yuxin Yuan, is now online in Nature Communications🎉 Many thanks to co-authors, Xiaoyu Li and @muzizimumu1, and to all
2
6
37
Delighted to collaborate with J. Hu and @HongyuZhao2 to show that genetic mechanisms leading to a high CAD polygenic risk score are heterogeneous, which may have implications for prevention https://t.co/dmOYWOsaWU
@PLOSCompBiol
2
9
40
Very excited to share our preprint led by @MGLevin @skoyamamd @jakob_woerner & with @damrauer assessing genome-wide pleiotropy of >1,000 clinical traits across ~1.7M individuals with nearly 30K locus-trait associations! https://t.co/hJChZPSYNm
@medrxivpreprint [1/3]
3
16
82
Excited to share our new manuscript on evaluating multi-ancestry GWAS methods using large-scale simulations & real data from UK Biobank (N≈324K) & All of Us (N≈207K)! My first thesis paper under the guidance of @GENES_PK & @AndrewHaoyu. Available at: https://t.co/xxAbJXn7zZ
1
6
30
Our new PRS paper is out! We introduce a method to create regularized ensemble PRS on GWAS sumstats. Individual-level data is no longer needed for sophisticated ensemble learning @ZijieZhao1996 @stphn_drn Paper📰: https://t.co/fUZD6TXzdj Software🧑💻: https://t.co/YIYhPAIDpY
2
21
57
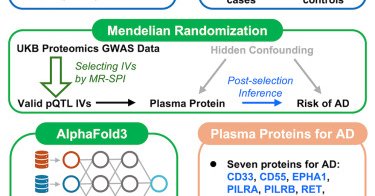
Excited to share our latest work in Cell Genomics! Using a novel MR method integrated with AlphaFold3, we identify Alzheimer’s-associated proteins with 3D structural changes. https://t.co/Q5lnLo7zqn
@BaccarelliAA @GaryWMiller3 @ColumbiaMSPH @CellGenomics
cell.com
Yao et al. develop a computational pipeline integrating a robust Mendelian randomization method with AlphaFold3 to select valid pQTL instruments, identify causal protein biomarkers, and predict 3D...
2
7
39
Thank you Pradeep for your support and advice for this project🙏😃 We’re thrilled to introduce DiscoDivas, a new method designed to generate Polygenic Risk Scores (PRS) for diverse espcially admixed populations 1/
Current polygenic risk scores require binning of ancestry groups but we know ancestry is a continuum. Our study led by @Yunfeng_Ruan introduces DiscoDivas, a new generalizable PRS framework across the ancestry spectrum https://t.co/MCduxNKpSg
@medrxivpreprint @PRSdiversity
2
16
38
Current polygenic risk scores require binning of ancestry groups but we know ancestry is a continuum. Our study led by @Yunfeng_Ruan introduces DiscoDivas, a new generalizable PRS framework across the ancestry spectrum https://t.co/MCduxNKpSg
@medrxivpreprint @PRSdiversity
2
23
84
#ASHG24 Interested in how to reliably use AI/ML to accelerate genetic discovery? Stop by poster 4151 this afternoon to chat!
2
8
19
Really appreciate the tremendous support from @XihongLin, Rayjean Hung, Chris Amos, Laura Bierut and All collaborators! The first author @Tony_Chen6 is currently on job market for faculty and postdoc positions. Feel free to reach out to him if you have opportunities!
0
0
0
Very excited our paper for incorporating PRSs into clinical practice for lung cancer and smoking cessation appears online today @eBioMedicine. This work is led by @Tony_Chen6 , co-mentored with @lishiunchen.
Our paper introducing a PRS-based translation roadmap for lung cancer care is now published on @eBioMedicine ! Wonderful collaboration with @AndrewHaoyu , @lishiunchen , ILCCO and GSCAN towards the clinical implementation of PRS. https://t.co/pLqPpEzj6k
1
0
15
Interested in PRS methods using continuous ancestries information? Check out our recent preprint: https://t.co/j10Z38fpvu. The work will also be presented as a ASHG poster at Board 4128T Nov 7th 2:30-4:30 by an excellent PhD student @Tony_Chen6! Feel free to stop by!!
biorxiv.org
Polygenic risk scores are widely used in disease risk stratification, but their accuracy varies across diverse populations. Recent methods large-scale leverage multi-ancestry data to improve accuracy...
Excited to share our preprint for SPLENDID, a new PRS method using continuous ancestry interactions and biobank-scale individual level data. Many thanks to @AndrewHaoyu , Rahul Mazumder, and @XihongLin for their valuable guidance on this work! https://t.co/vb2i6lRekA
0
3
12
Interested in uncovering how genetic regulation of TERT splicing influences cancer risk? Join me at the “Cancer Risk: Novel Genes and Mechanisms” session this Wednesday, Nov 6, at 8:45 a.m. in the Four Seasons Ballroom 4 during #ASHG24! @prokuninaolsson @theNCI
0
3
10
Excited to share our preprint, describing a method for heritability partitioning with GWAS sumstats that significantly improves upon S-LDSC Led by the fantastic Hui Li, and co-supervised by @XihongLin
#ASHG24 poster 4089F https://t.co/RHE6Es1Joi
medrxiv.org
Heritability enrichment analysis using data from Genome-Wide Association Studies (GWAS) is often used to understand the functional basis of genetic architecture. Stratified LD score regression...
2
38
117
Truly humbled to work with such an incredible team. Huge thanks to Jacob Williams, @Tony_Chen6 , Hua Xing, @wendyWongSW, Kai Yu, @GENES_PK, @xihaoli
0
0
1