Jae Lab
@TheJaeLab
Followers
480
Following
492
Media
4
Statuses
177
Functional genomics lab at the LMU Gene Center in Munich, Germany. -Posts by LJ
Joined February 2020
Don’t miss this great opportunity! Deadline is September 13.
🚀Wanna do your PhD at QMB? Apply NOW! Engage in interdisciplinary research that integrates experimental and theoretical approaches, or focus solely on theoretical work, and become a leader in the emerging field of quantitative, systems-oriented bioscience. https://t.co/af3syVPeiD
0
0
3
📣Only three weeks left to apply! Exciting projects with renowned scientists are available. Don’t miss your chance! https://t.co/af3syVPeiD
@TheJaeLab @karen_alim @PhysOfLifeLMU @hornung_lab @khmelinskaia @LFMilles @SchwilleLab @RaedlerLab @BenediktSabass
0
4
6
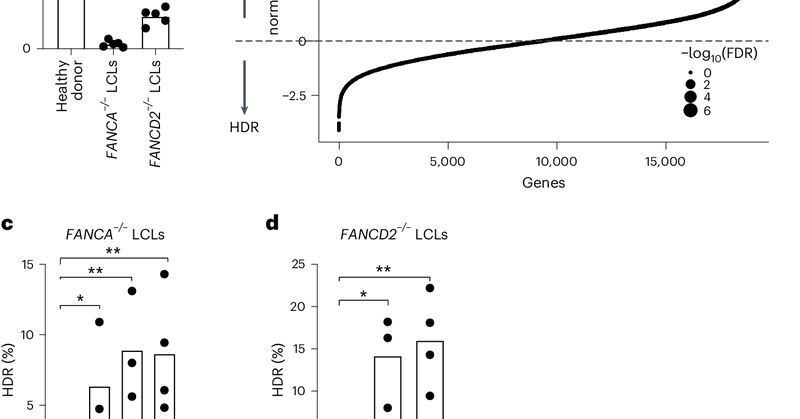
Having a hard time with #CRISPR HDR? Your cells might be fighting against you! TREX1 nuclease, high in some cells, eats up HDR donor templates. Check out our paper in @NatureBiotech for the story & ways to improve HDR. https://t.co/03GokR65LA With @karasuerman1 & @J_Maciejowski
nature.com
Nature Biotechnology - Homologous recombination in CRISPR–Cas9 genome editing is increased by blocking an exonuclease activity.
2
41
176
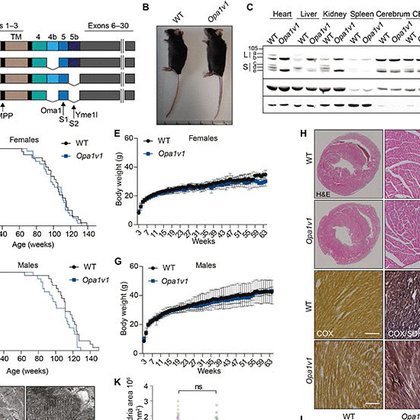
🥳🚨Paper Alert🚨🥳 Excited to present our latest work on #Opa1 in this newly published paper @ScienceAdvances by @sahola6 and @LPazurek. We present here two new mouse models with impaired Opa1 processing. 1/🧵 https://t.co/88eaOnzeO1
#mitochondria #mitochondrialdynamics @MPIAGE
science.org
Inhibition of Opa1 processing in mice allows normal development but exacerbates mitochondrial cardiomyopathy.
1
38
130
🚀Our celebratory symposium last Monday was enjoyed by all participants😀😆. Bavarian Minister of State Blume and LMU President Huber offered their congratulations. Patrick Cramer, President of the Max Planck Society, sent his greetings via video message.
1
3
13
There is a new study section @GBM_eV @JuniorGBM on Organelle Biology and I have just joined - exciting initiative by @winklhofer_lab & @Herrmann_lab and very relevant for our new @dfg_public Priority Program
3
5
31
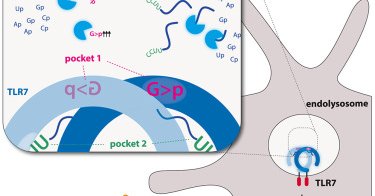
Excited to share our latest study in @ImmunityCP showing how PLD exonucleases together with RNase T2 process RNA upstream of TLR7. ✂️🧬 Kudos to @BeroutiMarleen for her fantastic work! https://t.co/nXXoCEcgmJ
cell.com
TLR7 is critical for recognizing RNA virus infection and initiating antiviral responses. Bérouti et al. demonstrate how RNase T2 and PLD exonucleases generate RNA fragments for TLR7 activation, thus...
7
27
108
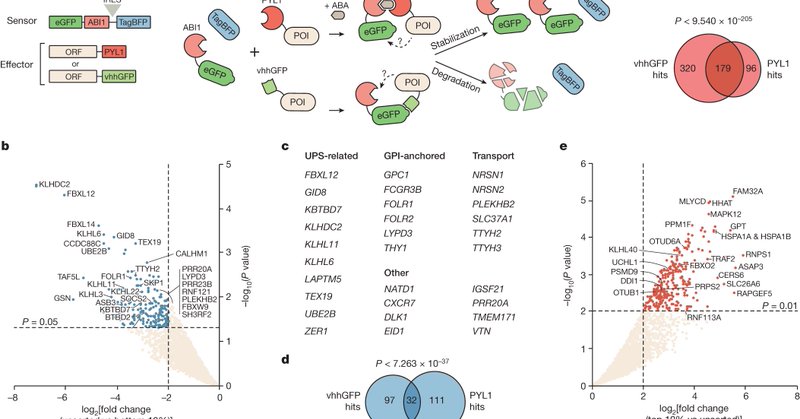
Our paper on functional discovery of protein degradation and stabilization effectors with proteome-scale induced proximity screens is out in its final version! 1/ https://t.co/vzNbJyR2rk
nature.com
Nature - A synthetic proteome-scale strategy enables the identification of a diverse range of human proteins that can induce the degradation or stabilization of a target protein in a...
27
145
557
Looking for a postdoc position to study epithelial tissue dynamics in Drosophila? New institute, interdisciplinary, international environment and very livable place @uni_muenster One week left to apply, see https://t.co/esjOrXhFiJ
We´re hiring! 2-year #postdoc position opening in my lab @uni_muenster to work on membrane trafficking and epithelial tissue dynamics in #Drosophila. For details, see https://t.co/qP5jZiNksB
@the_Node Please RT
3
16
26
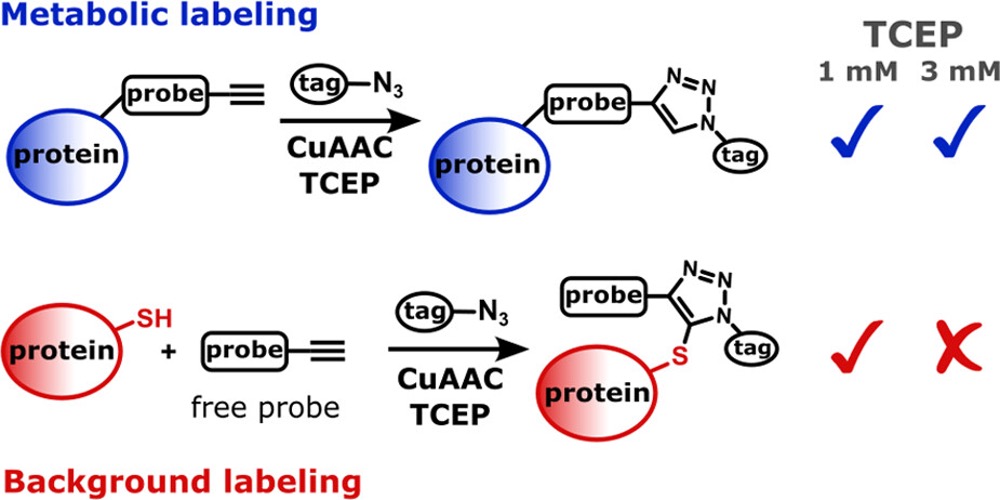
Let's do the right click! In chemical proteomics chemoselectivity of the CuAAC is crucial. Andreas Wiest in our group found that formation of the ubiquitous thiotriazole byproduct during the click reactions can be easily diminished: https://t.co/5uKlRx3n6i. Congratulations!
5
17
117
📣📣📣Great opportunity for ambitious junior researchers🫵 We are looking for an Independent Group Leader in Nucleic Acid Research🧬 https://t.co/fLEO8qyrFR Please RT🙏 #ApplyNow #Career
1
38
45
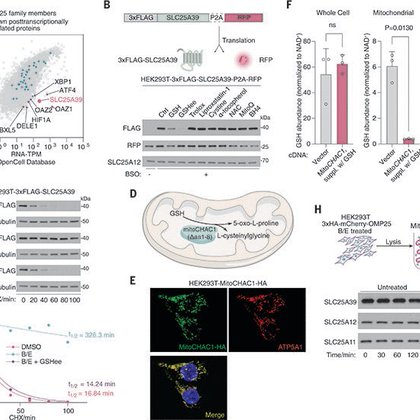
How does mitochondria sense and control their glutathione levels? Our new work @ScienceMagazine by a fantastic student @YuyangLiu1996 describing a redox metabolite sensing mechanism for mitochondria. @RockefellerUniv @TheMarkFdn
science.org
Blockage of an inner mitochondrial membrane protease stabilizes a glutathione transporter, enabling autoregulatory homeostasis.
46
157
656
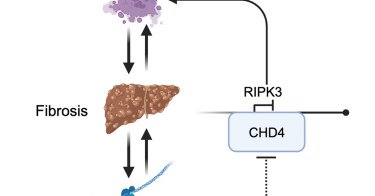
📢Don't miss this publication by the @hornung_lab Congratulations to all people involved👏🥳 β1 integrin signaling governs necroptosis via the chromatin-remodeling factor CHD4: Cell Reports https://t.co/0JzIWNfS9F
@LMU_Muenchen @237Crc @sfb_1403
cell.com
Sun et al. identify myofibroblasts as the major cell type expressing RIPK3 in bile-duct-ligation-induced liver fibrosis. Profibrotic β1 integrin signaling functions as the upstream driver of RIPK3...
0
4
11
Do cells care if their mitochondrial DNA is broken? Yes! Excited to share my PhD work showing that a fraction of cells with breaks in their mtDNA recover healthy mitochondria using the integrated stress response.
Online Now: Mitochondrial DNA breaks activate an integrated stress response to reestablish homeostasis https://t.co/WBy4ee8yGP
9
46
245
🥳🤩To our delight, Gonca Çetin @TheJaeLab has won a fellowship from the Peter und Traudl Engelhorn Stiftung. She will be supported for 2 years to investigate the integrated stress response in human cells. Congratulations, Gonca🎇🥂 https://t.co/lhurCun9oC
#Research #postdoc
0
1
14
I am so grateful to have been part of this! Congratulations to Drs Sekine and all co-authors! ☺️ @MolecularCell @TheJaeLab @AgingPitt #DELE1 #mitochondria
1
1
19
New paper out! We obtained conformational snapshots by using ATP analogues to reveal Mot1's special remodeling mechanism: gripping, bending and rotating upstream DNA to dislodge TBP from the TATA-box. @StephanWoike et al. https://t.co/E8MQzcmGxR
2
5
60
Have you ever wondered why some cells are highly susceptible to necroptosis, while others are not? Then take a look at our new preprint on how integrin signaling is a gatekeeper for RIPK3 expression. 1/5 https://t.co/9mltqgASy7
biorxiv.org
Fibrosis, characterized by sustained activation of myofibroblasts and excessive extracellular matrix (ECM) deposition, is known to be associated with chronic inflammation. RIPK3, the central kinase...
2
23
91
🥳🥂🎇Congratulations: Antje Boetius is the first winner of the Ernst-Ludwig Winnacker Award for enhancing the impact of science for the benefit of society @BayerFoundation #scienceforsociety @AWI_de @UniBremen @MarineMicrobio
2
6
40
🏃♀️🏃🏃♂️ Hurry up, only few days left to apply! 🔬🏔️🥨 @LMU_Muenchen @GeneCenter_LMU @ResearchGermany @mitojobs @mitowomen
Are you passionate about research? Can you feel the thrill of discovering secrets of nature no one has ever laid eyes on before? If you answered yes to these questions, the Functional Genomics division @TheJaeLab is the right place for you! https://t.co/KpXToGV9Pm
#phdpositions
0
2
5