Brian J. Abraham
@TheAbraLab
Followers
154
Following
0
Media
13
Statuses
45
Computational Biologist studying how genes are controlled in healthy and diseased cells
@StJudeResearch
Joined June 2014
I don't know that I've seen a project that ended so far and so much more important than the OG plan. From feelings that something is weird about CDK13 to battling the lack of reagents to separate it from CDK12 to tying stumbling polymerases to cancers. Big work; long time coming.
@leonard_zon @youngricka @AdelmanLab @NCIChanock @kbrowngenetics @TheAbraLab @dliu_ccb @bmart87 Normally, cells make gene expression mistakes, but luckily CDK13 triggers clean-up of “junk” RNAs from protein coding genes. We found that when CDK13 is mutated, these “junk” RNAs accumulate, driving more aggressive melanoma. Turns out, it is important to shred the trash!
0
0
4
St. Jude is seeking its next generation of graduate students. These students are exposed to the undersold breadth of research on campus and given tremendous resources to lead their own impactful research.
0
2
4
Our massive undertaking in finding vulnerabilities of pediatric cancers requires massive #bioinformatics talent. I'm excited to be a part of the team and am looking for scientists to join the effort. Posting forthcoming, but feel free to email your CV.
@broadinstitute @DanaFarberNews "Through #PedDep, we believe we can leapfrog barriers to rapidly identify therapeutic vulnerabilities in childhood cancer and translate those into targeted therapies in the clinic faster," says project co-lead Dr. Charles Roberts, St. Jude Comprehensive Cancer Center director.
0
2
11
Join the faculty of @StJudeResearch as we continue our major research/translational expansion. All levels, expansive scientific foci considered. More information here: https://t.co/L6ztYatukQ
0
5
6
Pleased that our review on cell state plasticity and core regulatory circuitry online! Check it out! Thanks to @CellRepMed, including @isabelgoldm and @DrSaraHam for their support!. @TheAbraLab @markw_zimmerman. #neuroblastoma #epigenetics
cell.com
Evidence has implic ated intratumoral heterogeneity as a driver of resistance to cancer therapies. Shendy et al. review how intratumoral heterogeneity is controlled by flexible transcriptional...
5
15
65
Analyzing sequencing data can be hard, especially if you don't have a seasoned analyst or heavy-duty hardware. So @Modupeore_A built a portable, modular, automated, cloud (and more) pipeline to do the most common ChIP-Seq/CUT&RUN-Seq analyses for about $20 per sample.
St. Jude coders built a portable, one-click platform called SEAseq to streamline analysis of chromatin pulldown sequencing data (ChIP-Seq and CUT&RUN-Seq). Read the @BMC_series Bioinformatics Paper to learn how. https://t.co/a9YDyiUeWm
@TheAbraLab
2
2
33
Deeply #thankful for strong, clever collaborators who like to tackle important problems, like this team. It made and makes sense for transcription factors that drive specific cancer types to be expressed only in those cancer types. @lawrensonlab built a way to find them.
0
1
4
Another foothold into drugging transcription shows that it matters in neuroblastomas. Strong collaboration among @ADurbinLab @JunQiLab @KStegmaier_DFCI and many other friends.
Scientists are sending neuroblastoma cells to the trash with a new compound that degrades EP300. https://t.co/FpMNOsvTaB
@CD_AACR @ADurbinLab
1
0
8
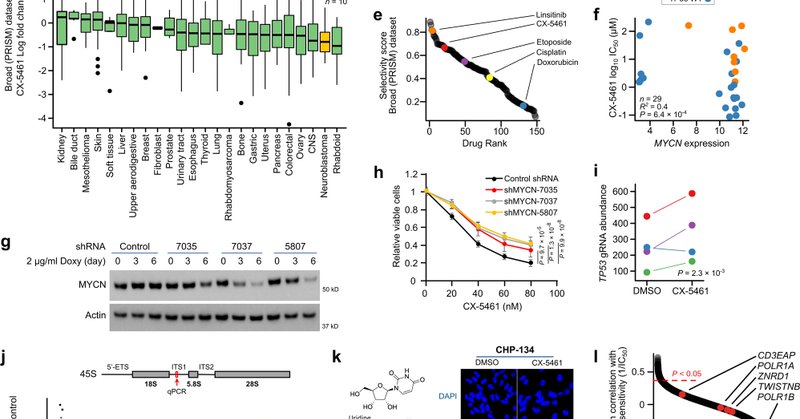
New paper out from our lab investigating the use of the putative RNA-POL I inhibitor CX-5461 in pediatric neuroblastoma. I believe this study represents a fantastic example of why the details of drug mechanism are deeply important for patient safety.
nature.com
Nature Communications - CX-5461 recently progressed through phase I clinical trial as a first-inhuman inhibitor of RNA-POL I. Here, the authors demonstrate that CX-5461 synergizes with...
1
15
32
Join us! St. Jude is recruiting multiple research faculty at the full, associate, and assistant levels in a major expansion of our basic and translational research programs. #cancerresearch #facultyjobs @stjuderesearch
https://t.co/L6ztYatukQ
0
3
4
Cancer patients with high-risk neuroblastoma often benefit from retinoic acid treatment. Read latest on how the drug works and what that means for the future of cancer treatment. https://t.co/v3WSrRvYDE
@ScienceAdvances @TheAbraLab
3
6
17
As a physician, scientist and president and CEO of St. Jude, Dr. James R. Downing speaks up for evidence-based medicine, science and the future of children. Read his new op-ed. https://t.co/wBfihCr9B2
#GetVaccinated #WearAMask @dailymemphian
1
44
76
Open staff #compbio #bioinformatics position in the Abraham Lab at @StJudeResearch. We study chromatin makeup and organization in a variety of cancer types. Apply here:
0
8
10
Join us July 22-23 for the virtual Bringing Chemistry to Medicine Symposium featuring thought leaders in areas spanning therapeutic regulation of transcription & chromatin, computational biology, and chemical biology. View speakers: https://t.co/4GGNgbYu98
#TranscriptionTherapy
0
9
9
To paraphrase Daria, there is no science that can't be improved with a group coffee.
Meet Whitehead Institute postdoc Ann Boija! Sometimes, Boija says, the best scientific ideas come not during long hours at the lab bench, but while sipping coffee with her fellow researchers. ☕ Read more about her experiences both in and out of the lab: https://t.co/CKo3I7mfLt
0
0
4
Join us for our first “The Science of Childhood Cancer” virtual lecture as Dr. Jinghui Zhang of St. Jude speaks on “Genomic Variants in Pediatric Cancer: Landscape, Precision Oncology, and Data Sharing Ecosystem” Thurs, Jan 28 at noon CT/5 pm CMT. https://t.co/fWyKfkmfJe
#SOCC21
0
15
30
A postdoctoral research associate position is available in the Computational Biology lab of @TheAbraLab focused on gene expression-regulation mechanisms in healthy and diseased cells. Learn more and share. https://t.co/Wgq6WdT3Pm
#postdoc
0
4
5
Last first author paper with Avik Choudhuri and Brian Abraham @TheAbraLab . From the best lab and the best mentor @leonard_zon Signaling matters!!
nature.com
Nature Genetics - Enhancer variants associated with red blood cell traits alter signaling transcription factor motifs, leading to changes in expression of genes that are upregulated during...
0
5
43
A great description of our new paper showing the interplay between genetics and the environment in human traits and disease. Missed signals add up over a lifetime and can manifest later as big deals.
discoveries.childrenshospital.org
New work introduces a previously unknown layer of human genetics: genetic variation in DNA enhancers' ability to respond to incoming signals.
0
1
5
If your DNA is the same your whole life, why do so many genetic diseases only show up in adulthood? We argue here that gene variants linked to diseases and traits affect cells' response to the environment, accruing damage over time.
1
3
14