Kurt Schmoller
@SchmollerLab
Followers
654
Following
2K
Media
11
Statuses
972
Munich, Bavaria
Joined July 2017
Finally, through collaborations, we show that the reporter works in budding yeast (with Francesco Padovani @frank_pado in Kurt Schmoller's lab @SchmollerLab) and in Drosophila (Mashiat again, with help from Abhi Sharma in Savraj Grewal's lab @SavrajGrewal).
1
2
6
It really does work amazingly!
mt-Kaede-HI-NESS @MitoMORPH is such an amazing reporter for mitochondrial DNA, and it works beautifully in budding yeast too! Check out the updated preprint to learn more: https://t.co/8iTvb2Bw77
@frank_pado @AlissaFinster
0
1
7
mt-Kaede-HI-NESS @MitoMORPH is such an amazing reporter for mitochondrial DNA, and it works beautifully in budding yeast too! Check out the updated preprint to learn more: https://t.co/8iTvb2Bw77
@frank_pado @AlissaFinster
I want to follow up on our recent preprint introducing a new genetic fluorescent mtDNA nucleoid reporter.
0
4
22
🚨 New segmentation model alert! 🚨🥳 You can finally use InstanSeg in our #BioImageAnalysis software #CellACDC! InstanSeg is a fast and multi-channel cell segmentation model by @petebankhead, Thibaut Goldsborough, & co. The multi-channel ability is very interesting!
1
25
142
Our preprint list is now up on FocalPlane. This week we focus on research in #bioimageanalysis. Drop us a message if we’re missing any preprints that you’ve been reading or writing! https://t.co/PVCnThfmRP
focalplane.biologists.com
Microscopy preprints - bioimage analysis - Discussions
0
14
47
Best end to the incredible PhD journey! So proud, and grateful to everyone involved along the way!
Cover & highlight #22 by @osman_lab N&V on senescence/p21 & the path to fibrosis Commentary on PIWI & RNAs and… CEP192 localises mitotic Aurora-A activity DELE1 maintains muscle proteostasis Carboxy-terminal polyglutamylation regulates Dishevelled LLPS … https://t.co/MRguhXAFSg
0
1
6
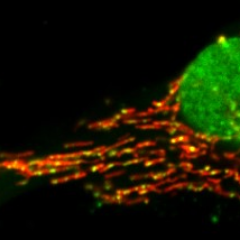
Seeing mitochondrial network on a lot of yeast cells on the cover of a prestigious journal makes me very happy 😎
Cover & highlight #22 by @osman_lab N&V on senescence/p21 & the path to fibrosis Commentary on PIWI & RNAs and… CEP192 localises mitotic Aurora-A activity DELE1 maintains muscle proteostasis Carboxy-terminal polyglutamylation regulates Dishevelled LLPS … https://t.co/MRguhXAFSg
0
1
5
Cell size matters, and it's a major regulator of cell function. @SchmollerLab reviews size control strategies of eukaryotic cells & the intricate link of cell size to intracellular biomolecular scaling, organelle homeostasis, cell cycle progression. 🔬 https://t.co/pyiQgMLrBm
0
9
23
The boss in action at the 6th Munich Yeast Meeting!✨️ Thanks to @SchmollerLab and @HamperlLab for organising such a great meeting! #Yeast #Mitochondria
0
3
15
To leverage the breathtaking progress of AI approaches for experimental biology, we need to bridge the gap between conceptual method development and application in real-life image analysis. We are happy to now be part of the great @bioimageio initiative with SpotMAX @frank_pado
🚀🎉SpotMAX is now a proud community partner of @bioimageio, the model zoo where you can find awesome pre-trained segmentation models! PS: You won't get the fireworks on the https://t.co/YXKKNvSP3l website but feel free to request them on the SpotMAX GUI 😎
1
4
15
While you could already use BioImage IO models in SpotMAX, by becoming a partner we commit to contribute with our pre-trained models, tutorials, etc. If you don't know what #SpotMAX is you can catch up on my previous Twitter thread here
SpotMAX, our software tool for analysing multi-dimensional microscopy data, is finally out! 🎉 And conveniently, I just presented it at the #I2K conference 😀 A multi-year-long effort of many great collaborators and users. But what can SpotMAX do? A small thread
0
1
5
SpotMAX: a generalist framework for multi-dimensional automatic spot detection and quantification Francesco Padovani et al doi: https://t.co/olHSjRuSg3
0
13
37
I am very happy to announce that I will be joining @HelmholtzMunich as a group leader from January 2025. Looking very much forward to work with my new colleagues on Ferroptosis and mitochondria #ERCStg @ERC_Research @Conrad_Lab Fabiana Perocchi @SchmollerLab @NatalieKrahmer
20
14
184
🎉One of our successful #epigenetics postdocs, @ATvardovskiy, is opening his own lab at @Penn! 🚨And he IS HIRING! Visit his lab website @PennEpiInst: https://t.co/MfSDd8KPYT & stay tuned for upcoming #academicpositions at all levels. Congratulations & best of luck, Andrey!
tvardovskiylab.org
📢 JOB ALERT: Tvardovskiy Lab at @PennEpiInst is hiring at all levels! Interested in chromatin biology, quantitative proteomics, or epigenetic modifications? Learn more about our research and apply at https://t.co/cz0RcZ6PRM Please RT
0
9
30
📢 JOB ALERT: Tvardovskiy Lab at @PennEpiInst is hiring at all levels! Interested in chromatin biology, quantitative proteomics, or epigenetic modifications? Learn more about our research and apply at https://t.co/cz0RcZ6PRM Please RT
0
68
220
SpotMAX: a generalist framework for multi-dimensional automatic spot detection and quantification https://t.co/v3ruuNEBUH
#biorxiv_cellbio
0
6
10
Oh, wow!Another #PrePrintAlert! Congratulations to @frank_pado, his supervisor @SchmollerLab & col. from IFE at Epigenetics @HelmholtzMunich!! A great #researchtool is now freely available to quantify spots from your #microscope #imaging. Read the fantastic 🧵from 1st author
You want to analyze your cells and need to quantify spots? But you don't want to develop your own pipeline from scratch or click on spots manually? @frank_pado and #spotmax have you covered! https://t.co/IDQyrzziBc
0
5
12
Congratulations @frank_pado! And huge thank you to our fantastic collaborators and the whole #spotmax team! @LabKoehler @daph_cabi @JetteLeng @xiaofengsu @NadaAlRefaie1 @YagyaChadha
0
2
5
You want to analyze your cells and need to quantify spots? But you don't want to develop your own pipeline from scratch or click on spots manually? @frank_pado and #spotmax have you covered! https://t.co/IDQyrzziBc
SpotMAX, our software tool for analysing multi-dimensional microscopy data, is finally out! 🎉 And conveniently, I just presented it at the #I2K conference 😀 A multi-year-long effort of many great collaborators and users. But what can SpotMAX do? A small thread
2
6
29
If you use SpotMAX, do not hesitate to contact us for any issues, questions, or feedback you might have! GitHub: https://t.co/JdpOtvehyl Documentation: https://t.co/GJQTeKrdLe Preprint: https://t.co/3bXpPGfsTo Ground-truth dataset:
github.com
SpotMAX: a generalist framework for multi-dimensional automatic spot detection and quantification - SchmollerLab/SpotMAX
1
1
13