Misha Kolmogorov
@MishaKolmogorov
Followers
2K
Following
675
Media
41
Statuses
2K
Tenure-track Stadtman Investigator at NIH/NCI @NCIResearchCtr. I like computational/cancer/metagenomics. The views are my own.
Bethesda, MD
Joined November 2017
A milestone for our lab! Here's a full text link:
Severus detects somatic structural variation and complex rearrangements in cancer genomes using long-read sequencing https://t.co/lh9zsE4Jdg
5
10
71
We are excited to announce our most recent preprint “Single-cell long-read sequencing of the experience-induced transcriptome” ( https://t.co/Qts0K63OmJ) led by @sheridan in collaboration with Richard Huganir. A few highlights:
biorxiv.org
Neural activity drives transcriptional events that are critical for learning. Activity-induced transcript isoform expression and alternative splicing are cell-type specific events typically obscured...
1
3
16
https://t.co/RfAn9FihsZ even after the DNA was repaired, the affected regions remained misfolded and showed reduced gene expression, and these changes were passed on to daughter cells. DNA damage thus leaves lasting marks on genome function, a phenomenon called “chromatin
science.org
Upon DNA breakage, a genomic locus undergoes alterations in three-dimensional chromatin architecture to facilitate signaling and repair. Although cells possess mechanisms to repair damaged DNA, it is...
0
3
8
Trio-barcoded @nanopre Adaptive Sampling (TBAS) to improve #RareDisease diagnostic at less than 1/2 the $$ & high cov: 76% solve rate across 13 trios inc. two corrections from prev. diagnosis. https://t.co/HbjvhTjTPn
@fuyvei96 @GREGoR_research @BCM_HGSC #Genetics #bioinformatics
0
7
30
Accurate somatic small variant discovery for multiple sequencing technologies with DeepSomatic https://t.co/i0nwOl1xBy
2
22
66
A telomere-to-telomere map of somatic mutation burden and functional impact in cancer https://t.co/xNsV0RTdf5
#biorxiv_genomic
1
8
24
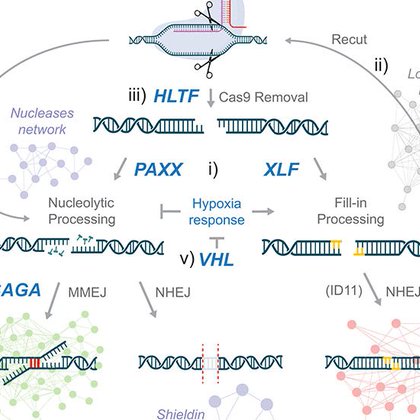
A comprehensive genetic catalog of human double-strand break repair | Science
science.org
The analysis of DNA sequence outcomes provides molecular insights into double-strand break (DSB) repair mechanisms. Using parallel in-pool profiling of Cas9-induced insertions and deletions (indels)...
1
20
72
Do you know ~60% of human SVs fall in ~1% of GRCh38? See our new preprint: https://t.co/ZdP2r83KI8 and the companion blog post on how we started this project and longdust: https://t.co/a9W6nXmslz. Work with @QianAlvinQin1
3
89
329
Preprint out for myloasm, our new nanopore / HiFi metagenome assembler! Nanopore's getting accurate, but 1. Can this lead to better metagenome assemblies? 2. How, algorithmically, to leverage them? with co-author Max Marin and supervised by Heng Li @lh3lh3
High-resolution metagenome assembly for modern long reads with myloasm https://t.co/lGIxJcirLX
#biorxiv_bioinfo
1
19
55
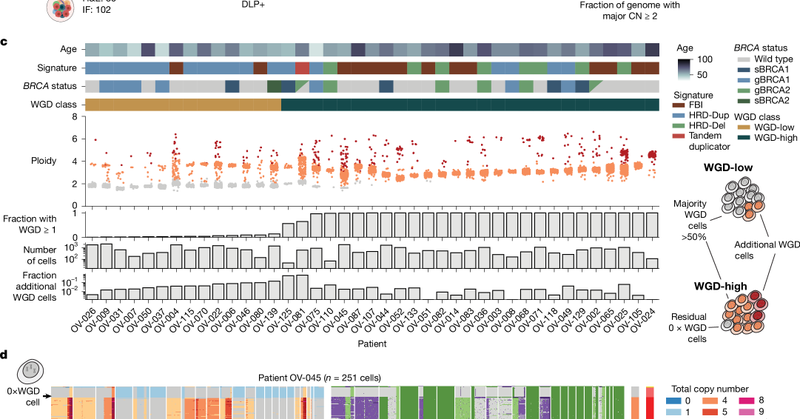
Whole-genome doubling (WGD) occurs in ~30% of solid tumors and is linked to poor outcomes. Is it a one-time event or an ongoing mutational process? In our new @Nature paper now in print, we show WGD continuously shapes tumor evolution and immune evasion. https://t.co/CLrFR2DpNB
nature.com
Nature - A single-cell sequencing study using more than 30,000 tumour genomes from human ovarian cancers shows that whole-genome doubling is an ongoing mutational process that drives tumour...
3
30
87
Our preprint on the challenges of benchmarking Structural Variants (SVs) is out in BioRxiv
biorxiv.org
Structural variants (SVs) are medium and large-scale genomic alterations that shape phenotypic diversity and disease risk. Numerous methods have been proposed for discovering SVs, however their...
3
9
20
Happy to share the latest work from the lab! We long-read sequenced single-cell derived melanoma subclones to reconstruct complete view of tumor evolution. With phylogeny, we can see parallel structural evolution and lineage-specific methylation.
biorxiv.org
Tumor evolution is driven by various mutational processes, ranging from single-nucleotide vari- ants (SNVs) to large structural variants (SVs) to dynamic shifts in DNA methylation. Current short-read...
0
6
35
Check out our study on the single cell derived sublines of a melanoma model: Long-read sequencing of single cell-derived melanoma subclones reveals divergent and parallel genomic and epigenomic evolutionary trajectories
biorxiv.org
Tumor evolution is driven by various mutational processes, ranging from single-nucleotide vari- ants (SNVs) to large structural variants (SVs) to dynamic shifts in DNA methylation. Current short-read...
0
2
10
Our newest computational method, ImmunoTyper2 for genotyping the complete germline immunoglobulin and T-cell receptor coding gene regions using WGS (short read) data is now out at Genome Research!
pubmed.ncbi.nlm.nih.gov
As we enter the age of personalized medicine, healthcare is increasingly focused on tailoring diagnoses and treatments based on patients' genetic and environmental circumstances. A critical component...
1
5
9
Clair3 now natively supports using GPU with CUDA or Apple Silicon to speed up calling in v1.2. using either a 4090, or a M3 Ultra, Clair3 can complete an ONT WGS 30x whole-genome variant calling in ~20min. A quick start is at 🔗 https://t.co/y7N7GTy9dt
1
12
43
Longdust, a new tool to identify highly repetitive STRs, VNTRs, satellite DNA and other low-complexity regions (LCRs). Similar to SDUST but for long regions. https://t.co/xUFgpmatJ5
github.com
Identify long STRs, VNTRs, satellite DNA and other low-complexity regions in a genome - lh3/longdust
2
70
200
Our Cell paper is online ( https://t.co/t5bOxlCN8q)! We present the CIDE framework, broadly applicable to cancer research and oncology. Also, we highlight AOAH as a promising new immunotherapy.
3
8
20
Identification of universally tumor lineage-informative methylation sites in colorectal cancer
academic.oup.com
AbstractMotivation. In the era of precision medicine, performing comparative analysis over diverse patient populations is a fundamental step toward tailori
0
2
1
Only 1 week left to register! This is great opportunity to meet and engage with your peers and faculty in algorithmic cancer biology, check out the speaker lineup!
SSACB 2025 rescheduled to Aug 18-22! We are inviting students and postdocs broadly interested in computational cancer to 2025 NCI Summer School on Algorithmic Cancer Biology. Please apply and spread the word:
0
1
2
Review: Structural variants in the 3D genome as drivers of disease https://t.co/GiI9gGAzFV (read free: https://t.co/UtJGMlLAxs) 🧬🖥️🧪
1
16
82
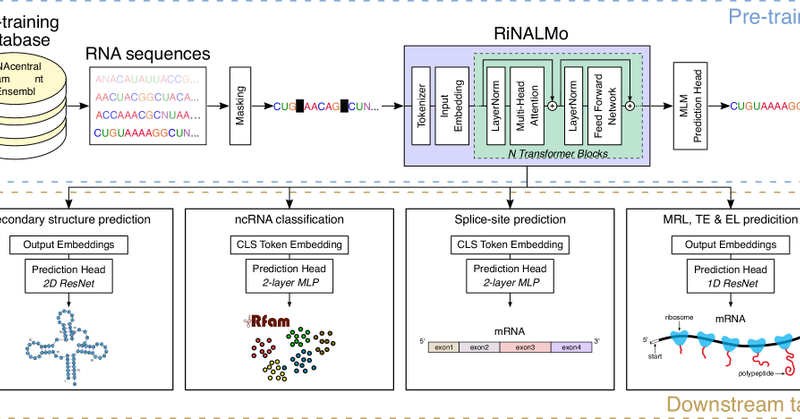
Our RiNALMo RNA language model has been published in Nat Comm https://t.co/2EXzExKdzK Great work by @RJPenic and @TinVlasic, with support and patience in teaching us RNA biology from Roland G Huber and @ywan_wan RiNALMo is already an SOTA as a benchmark for RNA LLMs.
nature.com
Nature Communications - RiNALMo, a large-scale RNA language model trained on non-coding RNA sequences, captures structural information and achieves state-of-the-art performance on multiple tasks,...
3
16
57