Rob Manguso
@MangusoLab
Followers
1K
Following
153
Media
89
Statuses
214
Official twitter of the Manguso Lab @MGHCancerCenter @BroadInstitute @harvardmed, studying cancer immunology using functional genomics and systems immunology
Cambridge, MA
Joined June 2019
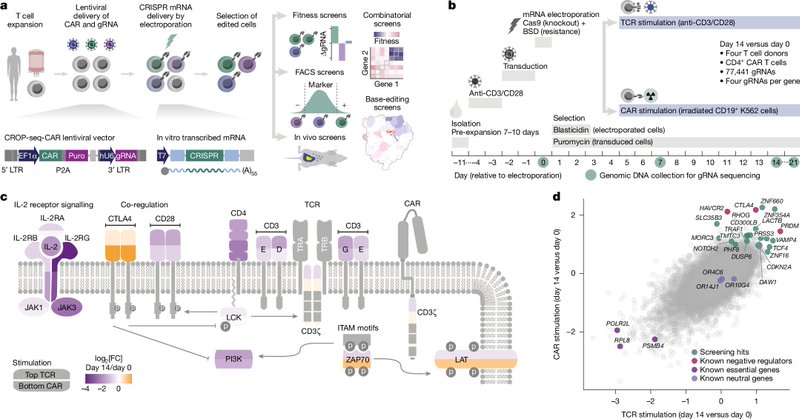
Super excited to share our latest work with @MarcelaMaus , out this week in @Nature. We show that in vivo CRISPR screens can identify genes that enhance the efficacy of CAR T cell therapy. Brief explainer below! @MGBResearchNews @broadinstitute
https://t.co/rUM3TAjkPu
6
15
88
Overall, we help inform the development of new immune-based therapies targeting HNSCC. At the advanced stage this is a deadly cancer that is poorly responsive to current immunotherapy approaches.
0
0
0
Interestingly, we find that deletion of Col17a1 alone (just 1 gene regulated by UCHL5) does enhance ICB response, although not to the same degree. Further, Col17a1 overexpression can reverse the ICB sensitivity seen in UCHL5 KO tumors.
1
0
0
In vivo we see that the MOC1es1 HNSCC tumors have very strong overall Col17a1 staining. As expected, UCHL5 KO tumors have lower Col17a1 staining.
1
0
0
We found this to be interesting because this collagen is expressed most strongly in HNSCC. In normal tissues it is found mostly in squamous epithelium. Deletion or overexpression of UCHL5 has the the expected effect on Col17a1, both in vitro and in tumors.
1
0
0
To gain mechanistic insight we performed RNAseq on KO and control cells. We find that UCHL5 KO cells have a downregulation of ECM and EMT related gene signatures, both in vitro and in vivo. Of note, we find strong downregulation of Collagen17A1.
1
0
0
Deletion of UCHL5 strongly enhances immune cell infiltration into the TME. We see a particularly strong increase in cytotoxic PRF1+ CD8+ T cells. We also find that CD8+ T cells are essential for the sensitivity to UCHL5 loss using antibody depletion studies.
1
0
0
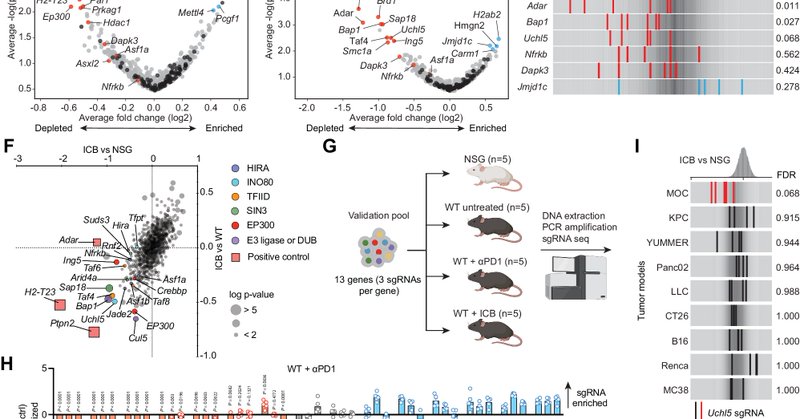
Deletion of UCHL5 in HNSCC tumor cells strongly enhances the response to ICB. We validate this in multiple mouse models of HNSCC. interestingly, this hit was not broadly seen in other types of cancer from our previous screens ( https://t.co/deVzK0WqgS). Possibly specific to HNSCC.
1
0
0
We performed in vivo screens using an epigenetic regulators-focused library in the MOC1esc1 model of HNSCC, treated with ICB, to find the regulators of ICB response. In addition to known regulators like H2-T23 (HLA-E), PTPN2 and ADAR, we find the de-ubiquitinating enzyme UCHL5.
1
0
0
Our latest work on IO target discovery is out this week. Amazing collaboration with @DrUppaluri and led by Cong Fu. We performed in vivo CRISPR screens in #HNSCC to identify targets to enhance ICB response. @MGHCancerCenter @broadinstitute @DanaFarber
https://t.co/pikSB4I2SE
nature.com
Nature Communications - UCHL5 is a deubiquitinating enzyme that cleaves Lys-48-linked polyubiquitin chains. Here, the authors discover through in-vivo CRISPR-Cas9 screens that Uchl5 is involved in...
2
7
30
Congrats to DrCongFu ! Wonderful to collaborate with @MangusoLab on in vivo CRISPR of epigenetic targets study in #HNSCC models @NatureComms @kathleenbyates @IOTNmoonshot Thanks also to @SilvioGutkind @rsaddawi !!
0
11
38
Congrats to Nelson Knudsen, @escobar_giulia, @FpkMd, and the entire team for the publication of their amazing work! And thanks for @MGHCancerCenter and @broadinstitute for the support!
0
0
1
Obviously this is a super exciting and fast moving space with all the great work coming out this week. We're happy to be helping to advance our understanding of therapeutic T cell biology, and there will be more to come!
1
0
1
BUT THERE IS MORE! Also want to highlight the preprint from @jccarnevale's lab from this week, showing that screens like this are possible at genome-scale in solid tumor models, and discovers regulators of GPCR signaling that block T cell infiltration! https://t.co/xgiBpfnvVF
biorxiv.org
Large-scale CRISPR screening in human T cells holds significant promise for identifying genetic modifications that can enhance cellular immunotherapy. However, many genetic regulators of T cell...
1
0
0
To this point, we were excited to see the great work from @BockLab that came out in the same issue of @Nature . Also a super impressive study and exactly the kind of work that will lead to a better understand of T cell biology in a variety of contexts. https://t.co/rZhLFvGh5O
nature.com
Nature - CELLFIE, a CRISPR platform for optimizing cell-based immunotherapies, identifies gene knockouts that enhance CAR T cell efficacy using in vitro and in vivo screens.
1
0
0
We suspect that this context specificity extends beyond simply the timepoint chosen or between in vitro and in vivo. It is likely that the profile of hits differs dramatically between tumor models. Ultimately as a field we need to do more of this work to unravel these differences
1
0
0
A few key messages: 1. KO of CDKN1B could enhance the efficacy of CAR T cells for multiple myeloma 2. In vivo screens can identify genes that promote CAR T cell expansion and persistence. 3. Context matters. We saw different hits in vitro, early in vivo, and late in vivo.
1
0
0
CDKN1B encodes the G1/S cell cycle checkpoint p27, and consistently when we delete it we see upregulation of G2M checkpoint genes. While it does have tumor suppressor roles, we showed that CDKN1B deletion doesnt transform T cells. We see no cytokine-independent growth.
1
0
0
When we look for CAR T cells in the bone marrow of tumor-bearing mice treated with either CDKN1B KO or control CAR T cells, we see a big expansion of the CDKN1B KOs. We also show complete tumor control in a second BCMA+ multiple myeloma model.
1
0
0
The differences between how the genes perform becomes more obvious using in vivo validations. Just as the screen predicts, PTPN2 deletion promotes tumor control early but it isnt sustained. RASA2 KO does not promote control. CDKN1B KO promotes long term tumor control in all mice!
1
0
0